RESULTS
I. MANUALLY ISOLATED POLYTENE CHROMOSOMES AND NUCLEAR ENVELOPES
1. LM and EM gross level observations on polytene chromosomes
During the salt dehistonization the chromosomes swelled considerably (in length and 2-3 fold in width) and most if not all band regions stayed clearly visible (Fig. l). Some bands seemed to get somewhat disrupted. The visibility of the bands was even more enhanced by incubation with RNase A at pH 5.2 (Fig. 2). This effect has been described with ethidium bromide stained chromosomes, which "looked denatured" after RNase A (Plagens, 1978).
Digestion of salt-treated chromosomes with DNase or sequentially with RNase and DNase abolished the visibility of the chromosomes and bands in fluorescence microscopy (data not shown). This was best shown when the staining of chromosomes was done with the DNA-specific fluorochrome DAPI in 2MS solution. After digestion with DNase no detectable fluorescence was seen after 10-30 min (or over night at 4oC), when inspected in the digestion buffer (with or without DAPI), indicating that no or undetectable amounts of DNA are present.
However, in phase-contrast optics (x40 dry objective) (Figs. 3, 4), residual chromosomes with bands could easily be seen. The bands appeared, after dehydration, UA staining, and air drying, as discs, toroids and arch-like structures in the same size range as the first expanded bands, with excellent resolution. Remains of nucleoli (Fig. 24) and puffs (dispersed) were also clearly distinguished.
Hyaluronidase (1 h), especially after salt dehistonization, was found to expand whole chromosomes even more than salt extraction alone. Further RNase and DNase treatment did not change the gross appearance of these expanded band scaffolds either (Fig. 5).
These residual chromosome (band) structures resistant to salt treatment and nucleases (at least DNase) will hereafter be called band scaffolds (cf. the scaffold of metaphase chromosomes in Adolph, Cheng and Laemmli, 1977).
2. Fine-level observations on polytene chromosomes after salt and salt-RNase treatment
2. 1. Salt-dehistonized chromosomes
Although band regions were clearly demarcated in the light microscopy of salt-treated chromosomes (without DNase), they looked bulky and aggregated in EM. Disappointingly little resolvable detail was evident even in stereo inspection (not shown). This resulted largely from the bulk of the unfolded dehistonized DNA that covers and obscures fine-structural details in both band and interband regions. This also involved the peripheral regions where DNA strands could be seen (Figs. 1 and 2) to extend out from the zone of "straps" (which stick out from the remaining chromosome; cf. Mullinger and Johnson, l979). Regarding the question of how DNA strands fold round these band regions, instructive new details which would add to those already described on other materials (Paulson and Laemmli, l977; McCready, Akrigg and Cook, l979; Mullinger and Johnson, l979, l980), were not found
The impression obtained was that the DNA was very much sheared in the washing steps. Only long entangled strands were seen but no well formed loops. The latter were typical in many preparations which served as controls for enzyme digestions and were kept (l h) in buffers (without DNase or with denatured DNase) after salt treatment. In these cases a great number of undisrupted long DNA loops could be followed to surround the peripheral strap zone of the remaining chromosome (Fig. 6). These DNA strands were smooth, showing that the extraction of histones (from their association with DNA) had been successful. Obviously, for most of the DNA (which almost floated free) it took a longer time than that provided to settle and attach firmly enough to mica in order not to be sheared by washing. This was favourable for successful DNA digestions because rapid attachment to the mica could be a hindrance to complete digestions.
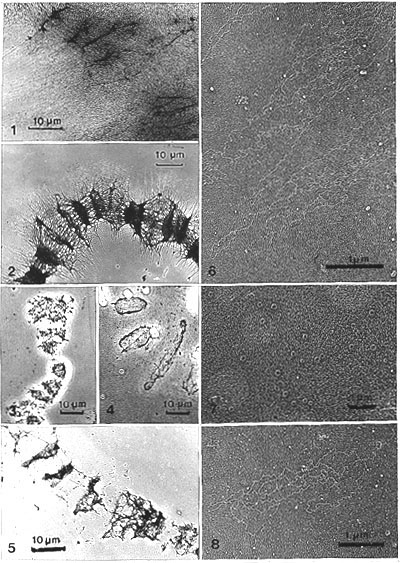
Fig. 1. Pilot experiment: Segment of a manually isolated Chironomus polytene chromosome, salt-dehistonized after whole-mounting onto a formvar-coated grid (made hydrophilic with cytochrome c). A large amount of decondensed dehistonized DNA surrounds the band regions. Bands are still distinct, yet the fine structure is hidden by the bulky decondensed DNA. Stained with uranyl acetate in methanol (x 1 200 ). Fig. 2. Segment of a chromosome, salt and RNase treated after attachment to mica. Bands appear contracted. The "strap zone" (in which "straps" protrude from the chromosome) is clearly visible. Pt-carbon replica (x 800). Figs. 3 and 4. Phase-contrast light micrographs of hand-isolated and whole-mounted chromosomes on mica. Segments of chromosomes, after treatments, UA staining, dehydration and air drying. Fig. 3. Salt and DNase treatments. Fig. 4. Salt, RNase and DNase treatments leave behind residual bands, "the band scaffolds." Note the concentration of residual material in the periphery, especially in Fig. 4, making the residual disc-like band scaffolds appear in phase contrast as ring and arch-like structures which match the size of the bands (x 800). Fig. 5. Segment of a chromosome. Sequential treatment with salt, hyaluronidase, RNase and DNase on mica. In gross appearance the band scaffolds treated with salt, RNase and DNase (without hyaluronidase) are similar. Pt-carbon replica (x 1 200). Fig. 6. Dehistonized DNA fibers protruding from salt-dehistonized chromosomes that were further incubated in control buffer (with denatured DNase for 1h). Long smooth loops settled down on the mica support. They originate somewhere in the strap zone. Pt-carbon replica (x 20 700). Fig. 7. Detail of Fig. 2, at higher magnification, from just outside the "strap region" (salt, RNase treatment). Many ring structures (diameter about 0.1 Ám) are visible. They resemble NPCs and are apparently embedded in undefined background material which could represent densely packed dehistonized DNA. Pt-carbon replica (x 9 700). Fig. 8. Material located further out from the strap zone (as in Fig. 6). Treatment as in Fig. 7. The undefined material of Fig. 7 now resolves into DNA loops loosely associated with the rings. Pt-carbon replica (x 16 000).
2. 2. Salt and RNase-treated chromosomes
The sparsity of unsheared DNA loops was also noticed in most preparations that were digested with RNase (no DNase) after salt extraction. The incubation time was short (15-30 min, which again is interpreted as not being long enough for DNA strands to settle and attach firmly) but long enough to elicit the "RNase effect" (Plagens, l978) in LM and to remove all digestable RNA.
After RNase treatment, however, there were indications of an uncovering of the structural nature of the scaffold: many ring-like assemblies, somewhat similar to NPC, although of variable size (Fig. 7), were observed in the peripheral (strap) region of the chromosome. These rings were only occasionally seen associated with DNA loops (sheared?) in the peripheral parts (Fig. 8), but in the strap region, closer to the chromosome, they were seen embedded in some unclear, slightly rough background material (Fig. 7). This material may be a layer of dehistonized uncoiled DNA which is too tightly packed to permit distinction of separate strands.
3. The NE scaffold: The framework of the nuclear envelope after salt and nuclease treatment
For interpretation of the structure of band scaffolds the comparison with similarly treated NEs was needed. After salt and detergent extraction of NE whole mounts, a nuclear pore-lamina fraction stays (Scheer, Kartenbeck, Trendelenburg, Stadler and Franke, 1976); and well preserved nuclear pore complexes (NPCs) are also seen in thin section studies using a similar extraction and nuclease posttreatment in bulk (Aaronson and Blobel, l975; Dwyer and Blobel, l976). NPCs should be distinguishable also in the carbon and rotary Pt-carbon replicas of whole mounts. Obvious NPCs and their annular subunits were found.
Seen in stereo pairs the manually isolated NEs of salivary gland cells (Fig. 9) appeared after salt treatment as collapsed and flattened spheres that do not change further after RNase and DNase treatment, both in light and electron microscopy (Fig. l0). Details in stereo pairs (Figs. l3-l5) showed that generally the NE is a layer of tightly packed small (25-30 nm in diameter) globular structures (which we have called globosomes, Engelhardt and Plagens, 1980a). They were often assembled around a cavity or a central hole (inner diameter 25-50 nm).
This compound organization (outer diameter 80-120 nm) of globules surrounding a central hole, definitely resembles the NPC with the peripheral globules representing annular subunits. Stereo observations also show cavities much larger than the central hole of the NPC. This can be interpreted as extended interpore spaces and/or artifactually expanded and stretched clusters of NPCs. After further digestion with RNase (not shown) or RNase and DNase (Figs. 13-15) these structures stayed apparently the same. The dense packing of pore complexes in NEs of salivary gland cells almost excluded their individual inspection.
Therefore, as auxiliary material, nuclear matrices from isolated rat liver nuclei (Berezney and Coffey, 1974) on mica (including a 30 min. hyaluronidase digestion) were analysed. In these preparations the NEs were less tightly packed with NPCs.
As expected the structures identified as the NPCs of the salivary gland cells can be seen as distinct entities in rat liver NEs (Figs. l2, l6), because they are more separated. Their characteristic annuli are very distinct and their subunits appear identical to the globular structures which nearly fill the salivary gland NEs.
It can be concluded that the NEs of salivary gland cells, after salt and extensive nuclease treatments, are composed in a space-filling manner of globular structures (25-30 nm in diameter). These globular structures appear to be similar to the annular subunits of the NPC.
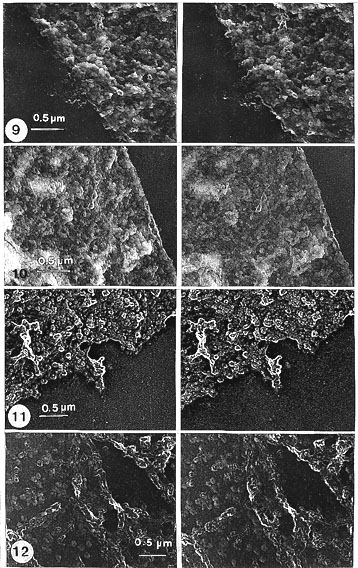
Fig. 9. Gross view of a sector (shown in stereo pairs) of a manually isolated NE from Chironomus salivary gland cells. Treated with salt only. Inspected in stereo, the NE appears to be composed of tightly packed globular structures (globosomes) arranged here and there around cavities of variable size, oriented in different directions. Notably, only one size class of cavity (inner diameter 25-50 nm) forms regularly arranged compound arrangements (outer diameter 80-120 nm), of globules surrounding a central hole, recognized as NPCs (encircled and seen in detail, in stereo, at higher magnification, in Fig. 13). Stereo pair of carbon replica (x 24 200).
Fig. 10. NE as in Fig. 9 but after the salt treatment digested with RNase and DNase. Neither the gross nor detailed appearance has changed (details of this at higher magnification are shown in Fig. 15). Stereo pair of carbon replica (x 24 200).
Fig. 11. NE treated as in Fig. 10 but Pt-carbon shadowed, for comparison. Stereo pair (x 20 000).
Fig. 12. Sector of a NE seen from isolated rat liver nuclei treated with salt, RNase, DNase and hyaluronidase (for comparison with those of Chironomus salivary gland NEs: Figs. 9-11). In contrast to the tightly packed NPCs in Chironomus NE, the NPCs are seen separated as distinct entities in rat liver NEs, with a characteristic annular arrangement of globular structures (globosomes). (Details in Fig. 16.) Stereo pair of carbon replica (x 20 000).
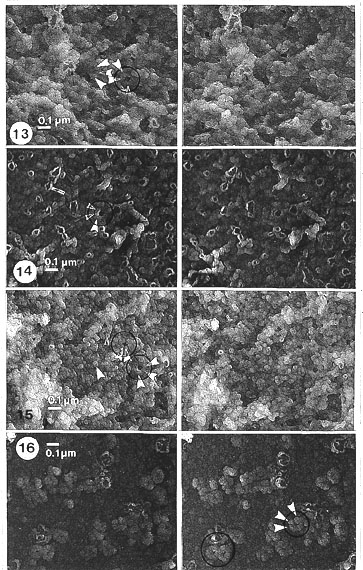
Fig. 13. Higher magnification of salt-treated NE. Fig. 13 is a detail of Fig. 9. Stereo inspection: details of the NPC (encircled) and its globular subunits (globosomes; white arrowheads). Most of the subunits are seen as being compact but some appear as ring-like or hollow, i.e. with a small, internal cavity, because in stereo inspection at high magnification a faint covering layer can be detected on the ring-like subunits (cyclosomes) (marked by black arrowheads). These subunits are of the same size class as the globular units, or sometimes fused to larger floppy rings (arrows). Stereo pair of carbon replica (x 45 000).
Fig. 14. Salt and RNase-treated NE. For some reason, the ring-like or hollow appearance of the subunits and their fusions are more frequent in this NE and particular treatment. Details marked as in Fig. 13. Stereo pair of carbon replica (x 45 000).
Fig. 15. Higher magnification of salt, RNase and DNase-treated NE. (Details of Fig. 10.) The appearance of the subunits (globosomes and cyclosomes) are unchanged by the treatments. Some details are even better shown than after salt alone, for example the subunits of the NPC are seen as more wedge-shaped. Details marked as in Fig. 13. Stereo pair of carbon replica (x 45 000).
Fig. 16. Detail of Fig. 12. Higher magnification of salt, RNase, DNase and hyaluronidase-treated NE of rat liver nuclei (compare with those of Chironomus NEs: Figs. 13-15). The NPCs seen as distinct separate entities with annular subunits clearly demarcated; some NPCs exhibit a wedge-shaped appearance. Details marked as in Fig. 13. Stereo pair of carbon replica (x 45 000).
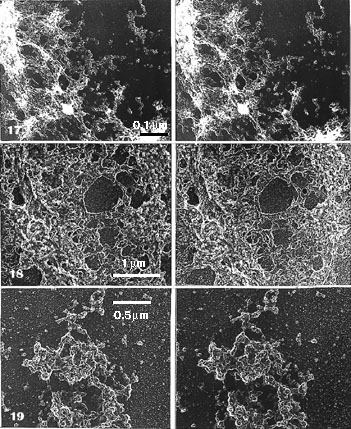
Fig. 17. Detail of a low magnification EM view of Fig. 3. (High-magnification details in Fig. 20.) Hand-isolated polytene chromosome (Chironomus) whole-mounted on mica: treated with salt and DNase-digested. A segment of band scaffold (lower left corner) and an interband region (upper right corner) presented (chromosome axis points from the lower left to upper right corner). The band scaffold appears in stereo inspection, at low magnification, as sponge-like, but in details similar to the NE, consisting of globular units (globosomes) and larger (stretched?) cavities than in NE; it is these cavities that give it the sponge-like appearance. As in the NE, one compound organization is prominent. It is of the same size range as the NPC, and in a similar fashion the globules are arranged around a rather regular-sized central hole (seen in more detail and encircled in Fig. 20). Note that in stereo inspection this compound ring structure, in the band scaffold, is more freely oriented, pointing in various directions, than the NPCs in NE. Because of this, it can be hard to detect without a stereo inspection. Stereo pair of carbon replica (x 8500).
Fig. 18. Detail of a low magnification EM view of Fig. 4. The same material as in Fig. 17, with the ring structures similarly oriented, treated with salt, RNase and DNase. The band scaffold is flattened and even more sponge-like, when viewed in stereo. Globular structures (globosomes) are very distinct, notably in the bulky part of the band region. The hollow or ring-like particles (cyclosomes) are also pronounced with their fusions seen as floppy rings. More details at higher magnification in Fig. 21 (band scaffold) and Fig. 22 (segment from the border of an interband region).
Stereo pair of carbon replica (x 16 000).
Fig. 19. Segment of a band scaffold. Salt, RNase and DNase treatment, as in Fig. 18, but Pt-carbon shadowing for comparison. The more compact band scaffold shows shiny rims of aggregated elements and outside, in the interband region, there are found single globular elements (globosomes). Note that the hollow or ring-like (cyclosome) appearance is not so demarcated in these Pt-carbon replicas as without Pt-carbon. Stereo pair of Pt-carbon replicas (x 24 200).
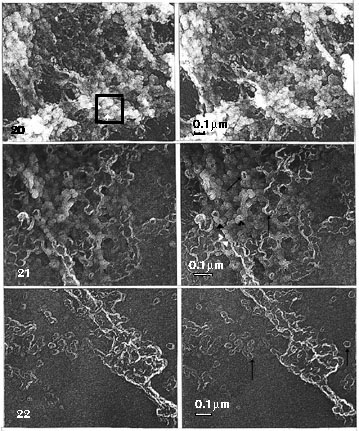
Fig. 20. Details at a higher magnification of Fig. 17, marked as in Fig. 13. Stereo pair of carbon replicas (x 35 000).
Fig. 21. and Fig. 22. Details at a higher magnification of Fig. 18, marked as in Fig. 13. Stereo pair of carbon replicas (x 45 000).
4. Fine structure of band scaffolds in polytene chromosomes
When only salt extraction was used, details of the residual bands could not be seen because of the effusion of the dehistonized DNA, as described. Further digestion with DNase (Figs. 17, 20) or with RNase and DNase (Figs. 18, 19, 20) resulted in residual bands which could look different in gross level appearance from the NE. In ultrastructural detail they were, however, indistinguishable from the salt-treated NEs.
Stereo inspection at higher magnification (Figs. 20, 21) showed that globular structures, of the same size as those found in the NE, formed the bulky part of band regions (the lower left corner of Figs. 17, 18) including the more dispersed material of interbands (the upper right corner of Figs. 17, 18), where these globules were seen separate or less clustered.
The globular particles had a distinct tendency to surround central holes just as in the NE where they form NPCs. The central holes could be collapsed, as in the NE, or somewhat expanded. Some holes, however, were greatly extended (similar to the presumed interpore spaces and/or artificially stretched and expanded clusters of pores in the NE), giving the whole scaffold a characteristic sponge-like appearance.
5. Hyaluronidase-digested scaffolds of bands and NEs
Digestion of the scaffolds obtained with salt and nucleases with hyaluronidase also yielded additional proof for the similarity of the particles in the framework of the bands and the NEs. Application of hyaluronidase (highly purified, no protease or DNase activity) to the bands and NE scaffolds produced a flow or "cascade" of particles extending out from all remaining band (Figs. 23, 27-33) and NE regions (Fig. 26). Regarding their origin, it is significant that the particles formed a steep concentration gradient extending as far as 20 mm from the main residual chromosome (Figs. 24, 25). Throughout the "cascade," besides single globular elements, their multiples (dimers trimers and oligomers) could easily be seen. The globular units in oligomers were clearly demarcated by gaps (Figs. 27-33).
General inspection showed a uniform size (diameter) for the globular units. In a sample of some 100 single monomers, 44% were 25-30 nm and the rest as follows: 22% 20-25 nm, 26% 30-35 nm, 7% 35-40 nm, 1% 40-45 nm. There seemed to be a tendency towards large size.
Although the particles looked rounded as viewed in stereo, preliminary photogrammetric (stereo) measurements indicated that they are flattened, being only about half as high (10-15 nm) as wide (25-30 nm).
The main oligomeric forms were linear or ring-like assemblies. Linear oligomers seldom exceeded 5-7 units. They were often slightly bent. Ring oligomers varied in diameter. Branched forms were infrequent (or interpreted as overlapping). There were also two and three-dimensional aggregates of different size classes. The units often looked compressed in the oligomeric forms (e.g. somewhat wedge-shaped in rings). Single units and terminal units of linear oligomers were rounded.
The band and NE regions, which in Pt-shadowed material remained after hyaluronidase treatment (Figs. 26, 27-30), displayed their subunit composition. As compared with band and NE scaffolds not treated with hyaluronidase (Figs. 11, 19), there were much clearer clefts between neighbouring globular substructures. It seems that hyaluronidase removed some material off the surfaces of these aggregated structures and demarcated them clearly.
In the sponge-like scaffold the structures that looked like robust fibres (or "cables" according to the nomenclature in a recent review by Earnshaw and Heck, 1988) clearly exhibited the same subunits in stereo. There were indications of some very faint fibrils at higher magnification (Fig. 30) in places where the granules were slightly separated. However, other robust fibres in the same figure broke abruptly without any underlying fibrous cores in the gaps.
Cascades were also produced in preparations of chromosomes that after salt treatment had been digested with hyaluronidase and then DNase (omitting RNase) (Fig. 32). Neither changing the order of digestions nor omitting RNase affected the production of cascades. Also, in Pt-shadowed preparations the appearance of the globular subunits was independent of RNase and the order of digestions. The particles had a shiny rim and looked spherical in stereo. Without stereo inspection the general impression of the particles could be that of small rings, because of the overwhelming density of this rim (Fig. 33), especially in the less electron-dense carbon replicas (Figs. 31a, b, c, 32).
The ring-like assemblies were detected throughout the cascades of band regions and of NE scaffolds. Their appearance was that of NPCs but the size was variable. The subunits of these ring-form assemblies could be distinguished but they looked more fused and did not always show the clear shiny rim as in the units of the linear forms (Figs. 27-30, 33).
Altogether, band and NE scaffolds responded to hyaluronidase indentically and in a very distinct way, suggesting that they were composed of very similar if not the same subunits. The evidence can be summarized as follows: (a) no differences could be detected in the particles or their assemblies under both categories; (b) in both cases the cascade formed a steep concentration gradient or zone round the remaining band regions and the NE; (c) neither the bands nor the NE were digested completely but a remainder was left (after 1 h digestion) and at the gross level there was little difference between hyaluronidase-digested scaffolds and undigested scaffolds except that the treated ones were usually more expanded and spread.
The application of hyaluronidase in different sequences (salt, RNase, DNase, hyaluronidase; salt, hyaluronidase, DNase; salt, hyaluronidase, RNase, DNase) always brought forth the same cascading of particles. Also, fine-structural details were independent of these changes in Pt-carbon replicas.
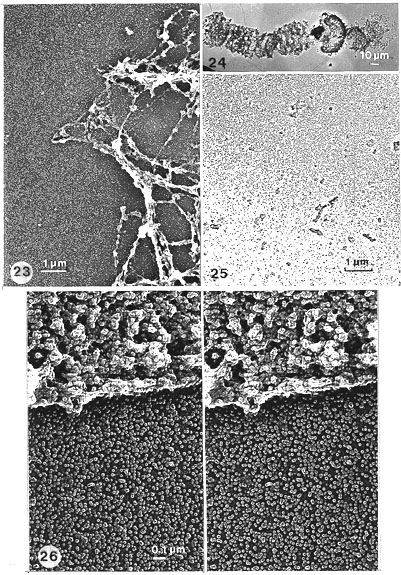
Fig. 23. A region including several band scaffolds after salt, RNase, DNase and hyaluronidase. Low-magnification EM view of the scaffold (compare with Fig. 5). At this magnification the cascade of globular particles (globosomes), in and outside of the band scaffolds, is evident. In the sponge-like scaffold rough fibrous structures are apparent and very pronounced (especially with RNase as here). Yet at a higher magnification of the same region, also the rough fibrous structures are seen, in stereo inspection, to be composed of similar globular elements (Figs. 27, 30). Pt-carbon replica (x 9000).
Fig. 24. A whole chromosome (including a residual nucleolus, arrow), after salt treatment and sequential digestion with DNase and hyaluronidase. Note that the bands are less contracted when RNase is not used. (Compare to Figs. 2, 4, 5, 23.) Pt-carbon replica. A low-magnification EM picture (x 330).
Fig. 25. The edge of the cascade which is about 20 mm from the chromosome scaffold. (Detail from lower edge of Fig. 24.) Note the steepness of the gradient of particles (globosomes). Pt-carbon replica (x 9000).
Fig. 26. A segment of a residual NE framework (scaffold) and an outside region with particles liberated in salt, RNase, DNase and hyaluronidase treatments. The cascade of particles (i.e. globosomes; also many oligomers) is evident. The shiny rim is pronounced, giving the particles a ring-like appearance. Stereo inspection, however, shows that the particles are globules, not rings. The NE is now more sponge-like, more like the band scaffold, i.e. with more and deeper cavities (especially when inspected in stereo) than the non-hyaluronidase-posttreated residual envelope (NE scaffold; compare with Figs. 9-11). The particular nature (globosomes) of the remaining NE is now pronounced. The band scaffolds (compare with Figs. 27-30), after hyaluronidase treatment, are similar. Stereo pair of Pt-carbon replica (x 35 000).
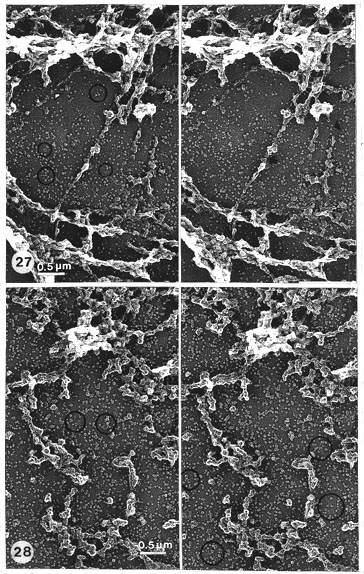
Fig. 27. Details of Fig. 23 at higher magnification and in stereo. Band scaffolds obtained by salt, RNase, DNase and hyaluronidase treatments. The background is covered by a cascade of particles. Many linear and ring-like (encircled) oligomers are seen. The residual scaffold is composed of particles similar to the cascade particles. There are rough fibres which are typical in RNase-treated material. The fibres are either continuous (thick arrow) or discontinuous (thick arrowhead). They are mainly composed of globular particles just as the rest of the more bulky regions of the band scaffolds. In places where the fibre is seemingly broken (small arrow) some core strands (undigested DNA?) are vaguely seen. (See Fig. 30 for more details at higher magnification.) Stereo pair of Pt-carbon replica (x 19 000).
Fig. 28. Band scaffolds (without RNase) obtained by salt, DNase and hyaluronidase (see Fig. 29 for more detail, at higher magnification). Compare with Figs. 23, 27, 30. Ring-like alignments encircled. Note that when RNase is omitted, no continuous fibres and no core strands are found. Stereo pair of Pt-carbon replica (x 19 000).
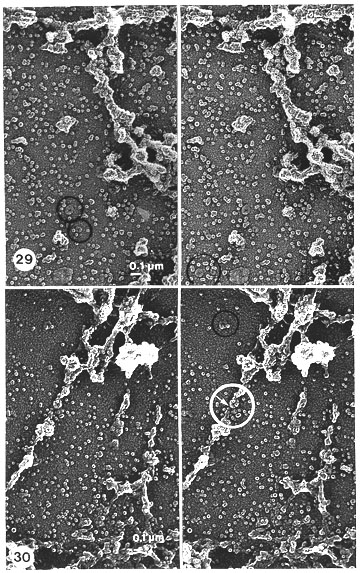
Fig. 29. Higher magnification of details in Fig. 28. All bulky remnants and minor clusters of the band scaffold reveal, in stereo inspection, that they are solely composed of the same globular particles (globosomes) as those distributed in the background including those that can be found in the residual NE after (i) salt only (Fig. 13), (ii) salt and nucleases (Figs. 14, 15), and after (iii) salt, nucleases and hyaluronidase (Figs. 16, 26). However, their appearance is more clear and demarcated, especially when compared to the band scaffolds obtained without hyaluronidase treatment (Figs. 19, 20, 21, 22). Note also that in the ring-like alignments (encircled) some particles have a wedge-shaped appearance, as in the NPC of the NE (Figs. 15, 16). No real, fibrous remnants can be detected even at high magnification. Stereo pair of Pt-carbon replica (x 36 500).
Fig. 30. Details of Figs. 23, and 27, at higher magnification, and viewed in stereo, to check the structural composition of the apparent rough fibrous structures seen in the sponge-like band scaffolds after the following treatments: salt, RNase, DNase and hyaluronidase. Stereo inspection reveals that the fibrous structures are composed of both smaller and larger clusters of the same globular particles (globosomes) that are visible everywhere in the background. The fibres appear to be just like some stretched parts of connections between adjacent, bulky band scaffolds (presumably because of the contraction effect that RNase digestion has had on the bands). However, some undefined glue-like material (undigested DNA?) can be seen stretched between the particles that seem to cause them to adhere together. This material is barely visible as very thin fibrils about 2-3 nm thick (indicated by a small white arrow, detected only within the white circled area).
Stereo pair of Pt-carbon replica (x 36 500).
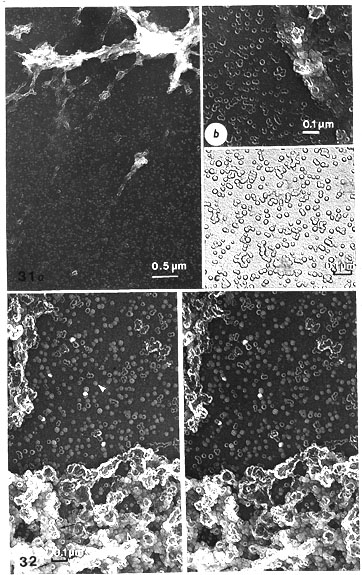
Fig. 31. a, b, c. Carbon replicas of hyaluronidase-digested band scaffolds to compare with the similar Pt-shadowed scaffolds of Figs. 23, 27, 30. Obtained by salt, RNase, DNase and hyaluronidase treatment.
a. A cascade of particles, which in this case appear as ring-like (cyclosomes), seen outside the band-scaffolds (x 19 000).
b. However, in the band-scaffold itself globular particles (globosomes) (marked with arrowhead) with the ring-like or hollow elements are evident, as in band-scaffolds treated without hyaluronidase (compare with Figs. 21, 22) (x 58 000).
c. Oligomers and aggregates are presented as seen in carbon replicas. The elementary ring-like particles are clearly recognized, although their inside borders are weakly seen, whereas the periphery of the oligomers and aggregates is very clear (x 62 000).
Fig. 32. Stereo pair of a carbon replica of a hyaluronidase-digested band scaffold (similar to Fig. 31, but without RNase digestion, to compare with the similar Pt-shadowed preparations of Figs. 28, 29) obtained by salt, DNase and hyaluronidase treatment. Even without stereo inspection a most apparent difference from Pt-shadowed material is obvious in that globular (globosomes) and ring-like particles (cyclosomes) occur side by side in the cascade produced by hyaluronidase. In Pt-carbon replicas, the heavier layer of contrast from Pt presumably hides the small difference in density between these two kinds of particle (x 54 000).

Fig. 33. A general view of particles and their alignments in a cascade outside a band scaffold after salt, RNase, DNase and hyaluronidase treatments. The particles are monomers, i.e. globular units (globosomes) and their compounded oligomers: linear and ring-form (encircled) alignments. Note the diversity in size of the ring-form assemblies. Unit particles can be distinguished in the rings, including some wedge-shaped examples (arrows), as earlier described in the NPCs of the NEs (compare with Figs. 15, 16). When some areas of the ring peripheries have less contrast, the unit particles are yet distinct because of their shiny rim. Pt-carbon replica (x 31 500).
6. DNA folding and scaffold elements
When nucleases were omitted and only hyaluronidase was used after the salt extraction of chromosomes (Fig. 34), the folding of DNA into many rosettes could be seen, often starting from ring-like cores. A considerable degree of unfolding permitted detection of the mode of folding. The shape of the cores was variable. Variants of (1) the ring-and-rosette (Figs. 34 a, b) were: (2) compact thick doughnuts of variable size, without DNA rosettes (Fig. 34 e) as were also (3) compact slightly bent rods with the same length as the doughnut periphery (Fig. 34 h); (4) thin straight structures with rosettes (Figs. 34 i, j). There were also aggregated clusters of such structures. Various degrees of disintegration of ring cores were seen; part or all of the ring could resolve into particles and the associated loops were then extended (Figs. 34 c, f, g). The extreme case is a ring of separate (eight?) particles without any connected DNA loops (Fig. 34 d).
These structures were the same rings that were already observed in salt and RNase-treated material, where the folding of DNA was less distinct (Figs. 7, 8). They were also encountered in the particle cascade of hyaluronidase-treated band scaffolds after both nucleases, as ring-form assemblies of variable size (Figs. 29, 30, 33). Interpretation of the cores, as inferred from the above morphologies, is as follows. Doughnuts and rods, being compact and smooth, hold the DNA tightly folded. The rod represents a linear conversion of the ring core. The rods also contain DNA which can unfold, so that the rods get correspondingly thinner
Typical, extensive cascades, in which monomers predominate (as described), were obtained from salt-treated polytene chromosomes only with hyaluronidase and nucleases (at least DNase). This is also the case with the NE (not shown).
As shown, the units were not always clearly demarcated when DNA was present in the core structures (of the NPC size), the reason for that being possibly that they are held together by DNA and stay hidden by the origins of DNA loops (compare Figs. 34 b, c and d).
In hyaluronidase-treated material, long DNA strands (some 5 mm) can be occasionally followed that seemed to connect different rosettes (Fig. 34 k). They probably broke up during the washing steps. In the normal course of treatment, the material expanded in salt, and more so in the digestion steps with hyaluronidase, leaving a cascade of scattered rosettes among some clumped material. The great abundance of separate rosettes, as described above, was seen especially when shrinkage of the chromosome bands during dehydration occured (obviously resulting from less freshly cleaved mica?), exposing the tightly attached rosettes, free from their bulky assemblies in the bands.
In places where the DNA loops were well spread (Fig. 34 b) some 8-20 loops per rosette could be counted with an average length of 0.5 mm per loop, which equals some 10 mm or 30 kb of DNA per rosette.
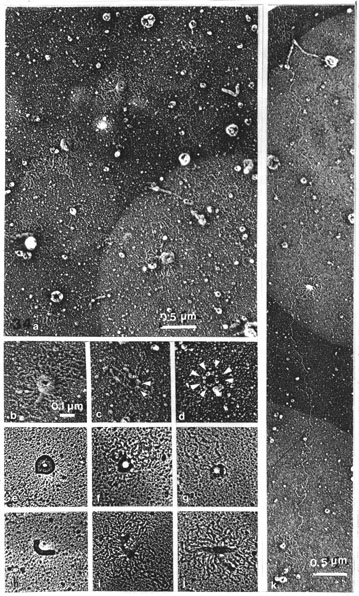
Fig. 34 a-k. The folding of DNA around scaffold elements after salt and hyaluronidase treatments
(omitting nucleases). Pt-carbon replicas.
a. General view. Abundance of ring-and-rosettes with doughnuts, rods and bent rods (x 24 000).
b-j. Details of the ring-and-rosette and its main variants: (a-d) and (k), negative prints;
e-j. positive prints of negatives (x 45 000).
b. Typical ring-and-rosette with many DNA loops; units of the ring are not distinguishable.
c, d, f and g. Stages of unravelling of the ring-and-rosette.
c. The connection of a DNA loop with globosomes; globosomes marked with arrowheads.
d. Separate (eight?) units can be distinguished, loops not visible.
e. Doughnut without unfolded DNA loops, with no distinguishable particles in the ring either.
h-j. Linear forms: compact rod without unfolded loops (h), unfolded loops (i and j).
k. A long, continuous DNA strand connecting rosettes of DNA loops (x 21 500).
Academic Dissertation 1988
Eukaryotic chromosome structure
[1]
[2]
[3]
[4]
[5]
[6]
[7]
[8]
[9]
[10]
[11]
Peter Engelhardt
Email: Peter.Engelhardt@Helsinki.Fi
Available at http://www.csc.fi/jpr/emt/engelhar/Doc/Diss-ResI.html










