Note
Click here to download the full example code
A spatial LDA beamformer¶
This example will demonstrate the LDA-beamformer spatial filtering approach on the MNE-Sample dataset. This dataset contains MEG recordings of a subject being presented with audio beeps on either the left or right side of the head.
The filter will attempt to isolate the timecourse of the auditory evoked potential.
This approach is further documented in van Treder et al. 2016 1.
First, some required Python modules and loading the data:
import mne
from posthoc import Beamformer
from matplotlib import pyplot as plt
path = mne.datasets.sample.data_path()
raw = mne.io.read_raw_fif(path + '/MEG/sample/sample_audvis_raw.fif',
preload=True)
events = mne.find_events(raw)
event_id = dict(left=1, right=2)
raw.pick_types(meg='grad')
raw.filter(None, 20)
raw, events = raw.resample(50, events=events)
epochs = mne.Epochs(raw, events, event_id, tmin=-0.2, tmax=0.5,
baseline=(-0.2, 0), preload=True)
left = epochs['left'].average()
right = epochs['right'].average()
contrast = mne.combine_evoked([left, right], [1, -1])
evokeds = [left, right, contrast]
Out:
C:\Users\wmvan\Miniconda3\lib\site-packages\sklearn\utils\deprecation.py:143: FutureWarning: The sklearn.metrics.scorer module is deprecated in version 0.22 and will be removed in version 0.24. The corresponding classes / functions should instead be imported from sklearn.metrics. Anything that cannot be imported from sklearn.metrics is now part of the private API.
warnings.warn(message, FutureWarning)
Opening raw data file C:\Users\wmvan\mne_data\MNE-sample-data/MEG/sample/sample_audvis_raw.fif...
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 25800 ... 192599 = 42.956 ... 320.670 secs
Ready.
Reading 0 ... 166799 = 0.000 ... 277.714 secs...
320 events found
Event IDs: [ 1 2 3 4 5 32]
Removing projector <Projection | PCA-v1, active : False, n_channels : 102>
Removing projector <Projection | PCA-v2, active : False, n_channels : 102>
Removing projector <Projection | PCA-v3, active : False, n_channels : 102>
Filtering raw data in 1 contiguous segment
Setting up low-pass filter at 20 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal lowpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Upper passband edge: 20.00 Hz
- Upper transition bandwidth: 5.00 Hz (-6 dB cutoff frequency: 22.50 Hz)
- Filter length: 397 samples (0.661 sec)
Not setting metadata
Not setting metadata
145 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
Loading data for 145 events and 36 original time points ...
0 bad epochs dropped
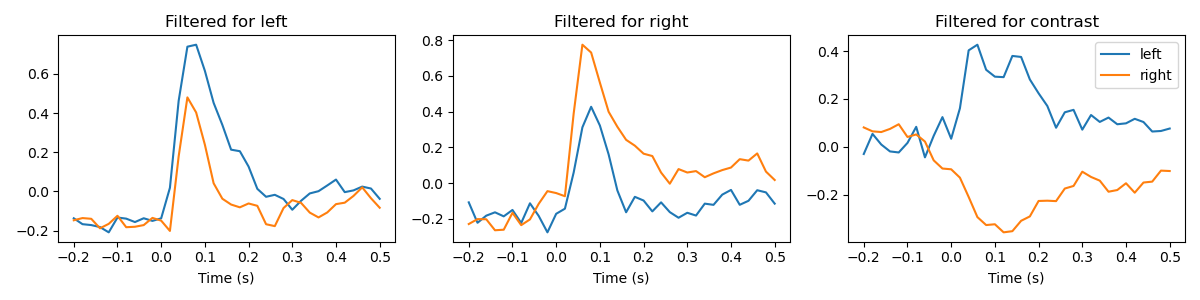
Now, we create some templates for the beamformer to use. A template of the potential that is evoked by the left beep, the right beep and a template that is the difference between the two conditions. We’ll try the beamformer with all templates and see the result.
template_left = left.copy().crop(0.05, 0.15).data.mean(axis=1)
template_right = right.copy().crop(0.05, 0.15).data.mean(axis=1)
template_contrast = template_left - template_right
templates = [template_left, template_right, template_contrast]
The posthoc package uses a scikit-learn style API. We must translate
the MNE-Python epochs object into scikit-learn style X and y
matrices.
def make_X(epochs):
"""Construct an n_samples x n_channels matrix from an mne.Epochs object."""
X = epochs.get_data().transpose(0, 2, 1).reshape(-1, epochs.info['nchan'])
return X
# Create data matrices in the scikit-learn format
X = make_X(epochs)
X_left = make_X(epochs['left'])
X_right = make_X(epochs['right'])
Design spatial filters using the different templates. We can give all the
templates at once to the posthoc.Beamformer object: it will
create separate filters for each template.
beamformer = Beamformer(templates).fit(X)
# Apply the filters to the left-beep and right-beep data repectively
X_left = beamformer.transform(X_left)
X_right = beamformer.transform(X_right)
# Plot the filtered evokeds
fig, axes = plt.subplots(1, 3, figsize=(12, 3))
for i, label in enumerate(['left', 'right', 'contrast']):
evoked_left = X_left[:, i].reshape(len(epochs['left']), -1).mean(axis=0)
evoked_right = X_right[:, i].reshape(len(epochs['right']), -1).mean(axis=0)
axes[i].plot(epochs.times, evoked_left)
axes[i].plot(epochs.times, evoked_right)
axes[i].set_xlabel('Time (s)')
axes[i].set_title('Filtered for %s' % label)
plt.legend(['left', 'right'])
fig.set_tight_layout(True)
References¶
- 1
Treder, M. S., Porbadnigk, A. K., Shahbazi Avarvand, F., Müller, K.-R., & Blankertz, B. (2016). The LDA beamformer: Optimal estimation of ERP source time series using linear discriminant analysis. NeuroImage, 129, 279–291. https://doi.org/10.1016/j.neuroimage.2016.01.019
Total running time of the script: ( 0 minutes 13.412 seconds)