Note
Click here to download the full example code
Plot sensor-level DSMs¶
This example demonstrates how to visualize representational dissimilarity matrices (DSMs) computed from EEG data. We will compute them using a spatial searchlight and plot the DSM computed for each searchlight-patch.
# Import required packages
import numpy as np
import mne
import mne_rsa
MNE-Python contains a build-in data loader for the kiloword dataset. We use it here to read it as 960 epochs. Each epoch represents the brain response to a single word, averaged across all the participants. For this example, we speed up the computation, at a cost of temporal precision, by downsampling the data from the original 250 Hz. to 100 Hz.
data_path = mne.datasets.kiloword.data_path(verbose=True)
epochs = mne.read_epochs(data_path / 'kword_metadata-epo.fif')
epochs = epochs.resample(100)
Reading C:\Users\wmvan\mne_data\MNE-kiloword-data\kword_metadata-epo.fif ...
Isotrak not found
Found the data of interest:
t = -100.00 ... 920.00 ms
0 CTF compensation matrices available
Adding metadata with 8 columns
960 matching events found
No baseline correction applied
0 projection items activated
We now compute DSMs using a spatial searchlight with a radius of 45 centimeters.
# This will create a generator for the DSMs
dsms = mne_rsa.dsm_epochs(
epochs, # The EEG data
dist_metric='correlation', # Metric to compute the EEG DSMs
spatial_radius=45, # Spatial radius of the searchlight patch
temporal_radius=None, # Perform only spatial searchlight
tmin=0.15, tmax=0.25, # To save time, only analyze this time interval
)
# Unpack the generator into a NumPy array so we can plot it
dsms = np.array(list(dsms))
# Visualize the DSMs.
mne_rsa.viz.plot_dsms_topo(dsms, epochs.info, cmap='magma')
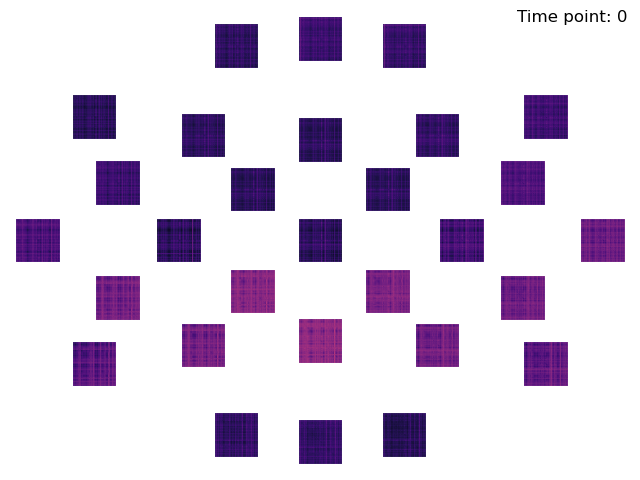
Creating spatial searchlight patches
<Figure size 640x480 with 29 Axes>
Total running time of the script: ( 0 minutes 4.120 seconds)