This example demonstrates how to perform cross-validation when computing dissimilarity
matrices (RDMs). When the data has repeated measurements of the same stimulus type,
cross-validation can be used to provide much more robust distance estimates between
stimulus types. Repeated measurements can for example be actual repetitions of the same
stimulus within the same recording, or recordings on multiple volunteers with the same
stimuli.
The dataset will be the kiloword dataset : approximately 1,000 words were presented
to 75 participants in a go/no-go lexical decision task while event-related potentials
(ERPs) were recorded.
This dataset as provided does not have repeated measurements of the same stimuli. To
illustrate cross-validation, we will treat words with the same number of letters as
being repeated measurements of the same stimulus type.
Authors
Marijn van Vliet <marijn.vanvliet@aalto.fi>
# Import required packages
import mne
import mne_rsa
MNE-Python contains a build-in data loader for the kiloword dataset. We use it here to
read it as 960 epochs. Each epoch represents the brain response to a single word,
averaged across all the participants. For this example, we speed up the computation,
at a cost of temporal precision, by downsampling the data from the original 250 Hz. to
100 Hz.
Reading C:\Users\wmvan\mne_data\MNE-kiloword-data\kword_metadata-epo.fif ...
Isotrak not found
Found the data of interest:
t = -100.00 ... 920.00 ms
0 CTF compensation matrices available
Adding metadata with 8 columns
960 matching events found
No baseline correction applied
0 projection items activated
The epochs object contains a .metadata field that contains information about
the 960 words that were used in the experiment. Let’s have a look at the metadata for
the 10 random words:
epochs.metadata.sample(10)
|
WORD |
Concreteness |
WordFrequency |
OrthographicDistance |
NumberOfLetters |
BigramFrequency |
ConsonantVowelProportion |
VisualComplexity |
| 635 |
hundred |
4.950000 |
3.552547 |
2.30 |
7.0 |
567.571429 |
0.714286 |
69.822284 |
| 462 |
emission |
4.950000 |
1.414973 |
2.35 |
8.0 |
344.750000 |
0.500000 |
66.092357 |
| 26 |
boil |
5.250000 |
2.100371 |
1.50 |
4.0 |
363.250000 |
0.500000 |
58.557164 |
| 938 |
friction |
4.400000 |
2.017033 |
2.25 |
8.0 |
567.625000 |
0.625000 |
52.045058 |
| 135 |
deceit |
2.700000 |
1.748188 |
2.15 |
6.0 |
603.333333 |
0.500000 |
65.893353 |
| 102 |
dinner |
5.700000 |
3.212720 |
1.75 |
6.0 |
1046.000000 |
0.666667 |
62.213401 |
| 526 |
brig |
5.866667 |
0.602060 |
1.65 |
4.0 |
532.500000 |
0.750000 |
65.524572 |
| 199 |
standing |
4.350000 |
3.283979 |
1.90 |
8.0 |
934.625000 |
0.750000 |
67.521514 |
| 513 |
trim |
4.150000 |
1.812913 |
1.50 |
4.0 |
597.500000 |
0.750000 |
56.238861 |
| 366 |
infant |
5.600000 |
2.571709 |
2.10 |
6.0 |
616.000000 |
0.666667 |
56.831681 |
The kiloword dataset as provided does not have repeated measurements of the same
stimuli. To illustrate cross-validation, we will treat words with the same number of
letters as being repeated measurements of the same stimulus type.
To denote which epochs are repetitions of the same stimulus, we create a list y
that contains integer labels for each epoch. Repetitions of the same stimulus have the
same label in the y list. This scheme is lifted from the machine learning
literature (and the Scikit-Learn API). In this example, we use the number of letters
in the stimulus words as the labels for the epochs.
y = epochs.metadata.NumberOfLetters.astype(int)
Many high-level functions in the MNE-RSA module can take the y list as a parameter
to enable cross-validation. Notably the functions for performing RSA and computing
RDMs. In this example, we will restrict the analysis to computing RDMs using a
spatio-temporal searchlight on the sensor-level data.
rdms = mne_rsa.rdm_epochs(
epochs, # The EEG data
y=y, # Set labels to enable cross validation
n_folds=5, # Number of folds to use during cross validation
dist_metric="sqeuclidean", # Distance metric to compute the RDMs
spatial_radius=0.45, # Spatial radius of the searchlight patch in meters.
temporal_radius=0.05, # Temporal radius of the searchlight path in seconds.
tmin=0.15,
tmax=0.25,
) # To save time, only analyze this time interval
Plotting the cross-validated RDMs
Creating spatio-temporal searchlight patches
<Figure size 640x480 with 29 Axes>
For performance reasons, the low-level functions of MNE-RSA do not take a y list
for cross-validation. Instead, they require the data to be already split into folds.
The create_folds() function can create these folds.
X = epochs.get_data()
y = epochs.metadata.NumberOfLetters.astype(int)
folds = mne_rsa.create_folds(X, y, n_folds=5)
rdm = mne_rsa.compute_rdm_cv(folds, metric="euclidean")
mne_rsa.plot_rdms(rdm)
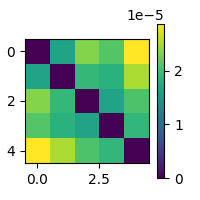
<Figure size 200x200 with 2 Axes>
Total running time of the script: (0 minutes 5.468 seconds)
Gallery generated by Sphinx-Gallery

