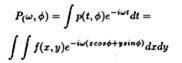CSC News (Vol. 6, No 4, p 14-20) December 1994
Electron Tomography of Chromosomes and Viruses
Peter Engelhardt1, Juha Ruokolainen2 and
André Dolenc3
1University of Helsinki, Department of
Virology
Email: Peter.Engelhardt@Helsinki.Fi
2Center for Scientific Computing (CSC)
3Helsinki University of Technology, Institute of
Industrial Automation.
Figs. 1. Human (HeLa, cell line) chromosome, low-pass
filtered to 20 nm.
Figs. 2. DNA-depleted human (HeLa, cell line) chromosome,
low-pass filtered to 20 nm.
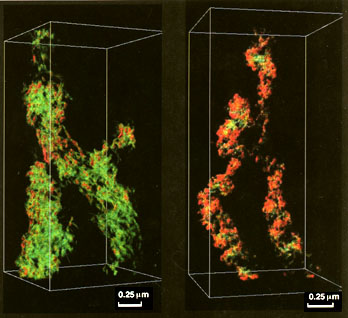
Fig. 3. Solid-3D-model of a tomography of a DNA-depleted
human (HeLa, cell line) chromosome (Fig. 2).
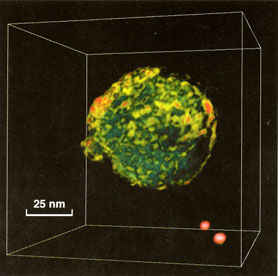
Fig. 4. Uukuniemi virus (Bunyaviridae), low-pass filtered
to 3 nm. Also two fiducial gold markers, colored red, can be seen in
the lower right corner. The virus is a close relative to Puumala
virus that causes nephropatia epidemica in Finland.
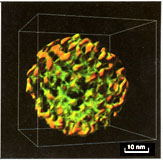
Fig. 5. Single-shelled particle of simian rotavirus
(Reoviridae), low-pass filtered to 3 nm. A human equivalent causes
diarrhea world wide.
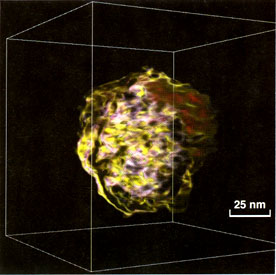
Fig. 6. Immature HIV, human immunodeficiency virus
(Retroviridae), low-pass filtered to 6 nm.
Introduction
An electron microscope is a structure projector of 3D-data. The
theoretical resolution of a transmission electron microscope (EM) is
at 100 kV, 0.002 nm, that is far below atomic resolution, deduced by
Prince Louis de Broglie (1926) from his description of the wavelength
of electrons (lamda= h/mv). The practical resolving power of most
modern electron microscope is, at best, 0.1 nm (1Å). The
picture of thick specimens is, however, superimposed, due to a high
depth of field, on photographic plates. In stereo-pairs the
resolution and the interpretation of structures are significantly
improved. In a more modern approach a complete 3-dimensional view of
the actual structures of microscopic matter is now available through
electron tomography [1].
In autumn of 1993 the first electron microscopy tomographies (EMT) of
chromosomes and viruses could be shown and used in Finland. This was
done at CSC by one of the authors (PE) who had obtained the
EMT-programs from docent Ulf Skoglund's laboratory at the Karolinska
Institute in Stockholm, after a half year training in using their
EMT-kit [2].
Our main efforts have been the further development of the performance
of the Swedish-EMT by introducing stereo-animation of the aligned
tilts at high resolution and sequences of the final volume around the
tilt axis with the capacities for interpolation, magnification and
measurement. All these features improve the visualization and
interpretation of the reconstructions significantly.
Steps of the 3D-reconstruction
EM-preparations
We will not go into all the details of how to prepare the material
for an electron microscopy study. This is beyond the scope of this
article. Many conventional EM-preparation techniques for both
thin-sections and for whole-mounts can be used. It is for the
individual investigator to decide how well the 3D-information is
preserved with various techniques. In our opinion the whole-mounts
are preferably critical-point-dried or alternatively preserved in
some other way, like sublimation from solidified t-butanol in vacuum
[3], to avoid shrinking. Also thin-sections are known to suffer from
uncompromised shrinking [4]. In addition, the whole object is not
necessary completely included in a single section that may or may not
be an advantage.
In our studies of chromosomes and viruses with the EMT we have found
that there is no consensus how to preserve chromosome [5] or virus
3D-structure. Though numerous whole-mount-techniques have been used
in chromosome research, only some, less conventional, seem to be more
suitable for 3D-studies [6].
However, the most well established whole-mount-techniques used to
study viruses, like the negative staining technique, have turned out
to be insufficient. According to our EMT studies, viruses are
flattened to 50 % with negative staining. The field is open for
searching new and better 3D-preservation-techniques for viruses.
Recently, after a systematic study of this problem, we finally found
some new and promising methods. The new methods reveal similar
delicate details like the negative staining method - used as a good
criterion - but avoids the shrinking problem. What we learn from
virus 3D-preservation studies can probably be used for other
biological material as well. This is partly the reason why we use
certain virus preparations as testing models in our EMT studies.
Moreover, some viruses have distinctive subunits or other decorations
in their shell that are of help as means of verifying successful
3D-reconstructions.
Tilt-series
The limiting resolution (d) for the tomographic reconstruction of
a spherical object diameter (D) from series of N projections equally
spaced between +90 and -90 degrees is given by Crowther-DeRosier-Klug
formula (for references see [5]):
d = pi*D/N
Obviously, increasing the number of projections improves the
resolution of the reconstruction. In practice, it will be difficult
to manually take tilt-series with smaller increment than 3 degrees,
which have become a standard for us, without observing electron
radiation damage to the specimen. A goniometer in a normal electron
microscope can only span from +60 to -60 degrees that results in 41
pictures (N = 60), with a 3 degree increment. This procedure takes 1
to 2 hours to complete and we have recognized practically no
radiation damage in magnification up to 58000x in e.g. whole-mounted
preparations of viruses. In magnification of 100000x serious
degradation of the specimen can be observed of whole-mounted
chromatin-fibers and viruses. Automating such procedures would
overcome this problem. Automation has recently been introduced [7-9]
but concerns modern electron microscopes that can be computer
controlled. However, no such microscopes are available in
Finland.
Scanning i.e. digitalization of the pictures
Lower magnifications of specimens are preferable over very high
magnifications not only because of danger for radiation damage to the
specimen but also because of focusing problems. At very high tilts
only the middle part along the tilt-axis will be in focus, the rest
will either be over- or under-focused. This is because of the limit
in depth of focus of ordinary (100 kV) electron microscopes.
The lower magnification can partly be compensated by scanning the
pictures with smaller raster size as the resolution of electron
microscope pictures is very high even at low magnification. The
flatbed scanner (Umax, UC630) at CSC has a maximum raster size of
about 50 mm that corresponds to a 1 nm size of the pixel at a 50000x
magnification. This usually exceeds the resolution obtained for a
reconstruction according to Crowther-DeRosier-Klug formula (above).
Nevertheless, there is a rule of thumb that says that the size of the
pixel should exceed the visible resolution at least 4x.
With the above settings we cannot presumably get better resolution
than about 4 nm for a reconstruction of a chromatin fiber or virus,
for instance. Apparently we will not see e.g. a DNA strand, diameter
of 2.5 nm.
A simple solution, but a laborious roundabout to secure a better
resolution in the scanning procedure, is to make prints. with e.g. 3x
magnification of the (41) negatives. We have recently tested this in
tomographies of viruses with excellent results and finished with an
improved reconstruction that could be low-pass filtered down to 3
nm.
Tomography
After the scanning of micrographs the resulting pictures must be
aligned with respect to one another. Scaling, rotations, and
translation of origin have all to be checked because of the
variations in them from one picture to another introduced by the
manual microscopy and scanning. This gives a set of over determined
nonlinear equations that can be solved, for example, by conjugate
gradient method. The aligning is done using fiducial gold particles
placed on the grid before the microscopy. The particle coordinates in
each picture must be picked interactively.
Having completed the alignment, the pictures are ready for the actual
reconstruction. The reconstruction can be done in 2D-slices which are
placed on top of each other to form the final volume. The usual
reconstruction method, which we have implemented ourselves, and which
the Swedish-EMT kit uses, is the weighted back-projection method (for
references see [1]).
The weighted back-projection is done by first Fourier transforming a
line from each projection (i.e. the micrographs) corresponding to
specific z-coordinate (along the tilt axis), weighting them by |w|,
(w being the spatial frequency), inverse transforming, and
backprojecting (a point in the projection with tilt Ø
corresponds to points (x, y) for which t = x cosØ + y
sinØ , t being the coordinate along the line in the
projection).
The method is given by the Projection Slice Theorem which says that
the Fourier transform of a projection corresponds to a radial line in
the projected objects Fourier transform:
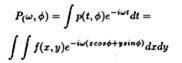
which gives, in polar coordinates
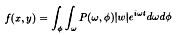
where t = xcosØ + ysinØ , and p(t, Ø ) the
family of projections (the Radon transform) of f(x,y)
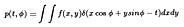
Presumably better reconstructions could be achieved using e.g.,
maximum entropy method, or reconstructing from estimated orthogonal
moments of the images.
Visualization of the 3D-volume
There are now several ways of rendering the 3D-volume:
BOB (stands for Bricks Of Bytes): The program is freely available
from Graphics and Visualization Laboratory of the University of
Minnesota Army High Performance Computing Research Center. BOB is an
interactive volume rendered for Silicon Graphics machines that uses
alpha blending.
The reconstructions are visualized in BOB as translucent objects
composed of voxels of different densities. In gray-scale the volume
is practically indistinguishable from the original EM pictures. With
BOB every voxel can be visualized in different translucent colors and
intensities, which clearly improves the visualization furthermore.
BOB uses also stereo-visualization of the volumes and many more
features of special advantage for the 3D rendering of very big
volumes.
FUNCS is the program we use for making promulgations of the volumes
with finest resolution. This program has been developed by one of the
authors (JR). The advantages in FUNCS are e.g. the incorporation of
lights and potentiality for interpolation to visualize the smallest
details. For big volumes all these features can be somewhat heavy to
be used interactively. However, the interpolated volumes can be
visualized smoothly by generating high quality animations in
jpeg-format around e.g. the tilting axis. The animations are run by
JPEGANIM that shows the jpeg-images produced in sequence and in
stereo. The animations can further be interactively magnified, moved
to different regions and details can be measured with scales at high
accuracy. FUNCS can produce animations also with the analogy colors
(red-green or -blue) for machine independent stereo-viewing [10].
We use the JPEGANIM program also for visualization of the original
tilt-series after the alignment procedure. With the animations we do
not only judge the quality of the alignment but also of the
3D-reconstruction as the original EM-tilt-series can be visualized
successively in stereo at high magnification as earlier reported
[10].
Hard copies i.e. solid-3D-models through 3D CAD
It is common nowadays to obtain a hard copy of a picture using a
computer. Given a three dimensional model, obtaining the
corresponding physical realization is, usually, very difficult or
even impossible. The geometrical complexity, for instance, in
3D-reconstructions of chromosomes is impossible to reproduce using
conventional manufacturing technologies such as numerically
controlled milling machines. Recent technological developments,
though, have made it possible to manufacture physical objects of
arbitrary complexity in mere hours [11]. These new manufacturing
processes are commonly referred to as "Rapid Prototyping
Technologies, RPT for short. A physical model can overcome some of
the shortcomings of computer generated images. Using this new
technology, we have manufactured a 3D-solid model of a reconstructed
chromosome. The reconstruction of the chromosome had to be converted
to a suitable representation. It was required, among other things,
that the various parts of the chromosome had to be attached by solid
bars in order to maintain their spatial relationship. The
manufacturing took place at SINTEF-SI (Norway) using a SOLIDER 5600.
We are also planning to manufacture a solid 3D-model of a
reconstructed virus.
3D-reconstructions
Chromosomes
We have made several reconstructions of eukaryotic chromosomes
that include metaphase chromosomes of a human cancer cell line (HeLa)
and Chinese hamster ovary (CHO) cells. Metaphase chromosomes are
isolated according to cytogenetical methods modified for electron
microscopy [6, 12]. The electron microscopy preparation contains
about 80-90% of pure metaphase chromosomes. A necessary amount and
size (5, 10, 15 or 40 nm) of fiducial-gold-markers are usually
included before chromosomes are applied to
plastic-coated-Ni-grids.
We take picture-tilt-series of whole-mounted chromosomes with
increments of 3 degree from 0 to + 60 degrees at 7000-20000x
magnification. At higher magnification e.g. 100000x, the chromatin
fibers of whole-mounted chromosomes are severely degraded by
radiation before the series is completed. As mentioned earlier, only
automation can effectively solve the problem.
The higher order folding of the DNA is an ultimate problem in
eukaryotic chromosome research. Nucleus of e.g. a human cell contains
about 2 m of 2.5-nm-thick DNA which in metaphase is divided into 46
chromosomes with a total length of about 200 mm i.e. a packing ratio
of 1:104. It has been clarified that the first order of packing is an
11-nm thick string of nucleosomes. In this "beads-on-a-string" form
of the chromatin, the DNA is wound 2 times around each nucleosome
composed of histone proteins. The second order packing of the
nucleosomes into 30 nm-chromatin fibers, which are the fibers usually
seen in EM-preparations of nuclei and metaphase chromosomes, has been
a controversial problem [5]. Electron tomography has also been used
in these studies [5]. The packing of DNA into 30-nm fibers gives a
packing ratio of 1:40; there is, however, still a higher order
packing of 1:250 to reach the metaphase level. The
higher-order-folding of the 30-nm chromatin fiber in metaphase
chromosomes and in interphase nuclei has been a central topic in
electron tomography [5]. Suitable preparative conditions are also not
agreed on [5]. Because of some unknown reason, specially the
chromosome-coiling is difficult to preserve with conventional
EM-preparative techniques [6, 13].
The preparative methods we have used [6] preserves the higher order
structure of chromosomes e.g. chromosome coiling as well as the 30-nm
chromatin fibers as shown in our 3D-reconstructions of chromosomes
(Fig. 1). Though, it is clear from our reconstructions that the
folding of the 30-nm fiber is much too complicated in eukaryotic
chromosomes to be easily followed in 3D. A solution to this problem
could be through some algorithm that could possibly resolve or track
the folding; though earlier efforts have been less prosperous [5]. In
this respect a better solution to the problem can presumably be found
from e.g. yeast chromosomes where the amount of DNA per chromosome is
much smaller.
Enzymatic digestion with DNase reveals a scaffolding structure of the
chromosomes [12] (for earlier references see [6] ). The
3D-Reconstruction of the chromosome scaffold shows a complicated
structure (Fig. 2). DNA-fluorescence cannot be detected with any
DNA-sensitive fluorochromes e.g. DAPI in the DNase-digested
chromosomes. Nevertheless, ordinary (Fig. 1) and control preparations
show bright DNA-staining. The spiral structure of chromosomes i.e.
chromosome coiling is often believed to be due to condensation of
chromatin fibers [3]. We can further show in the 3D-reconstruction
that the spiral structure is clearly maintained in DNA-depleted
chromosomes and rather due to the innate structure of the scaffolding
elements (for more details see [6] ).
The coiled structure of the chromosome scaffold is also very evident
in the 3D-solid-model (Fig. 3). It is also clear in the solid model
that in stretched regions, close to the centromere, we can
distinguish a 30-nm thick scaffolding-fiber that seems to coil into
100-150 nm in diameter macro-coils or regularly arranged rings as
earlier suggested [6]. In favorable situations e.g. the stretched
region, the scaffolding fiber seems also to reveal 30-nm thick
particles [6, 12, 14].
Viruses
Our 3D-reconstruction studies have been of RNA viruses. From other
studies following data can be found:
Uukuniemi virus (Bunyaviridae): A spherical, enveloped i.e. membrane
bound virus (90-100 nm in diameter) with an inner core i.e. capsid of
three 2-3 nm thick coiled, possibly helical, circular nucleoproteins.
The genome consists of three segments of single-stranded (ss) RNA
molecules of negative sense. The envelope is characterized by
glycoprotein knobs (10-12 nm in diameter) attached to the surface of
the lipid membrane. These are revealed with negative staining or by
freeze-etching techniques [15].
Rota virus (Reoviridae): 3D reconstructions of rotavirus particles in
cryo-electron-microscopy have revealed the structure at 4 nm [16].
Icosahedral, non-enveloped, double-shelled virions (outer 75-80 nm,
inner 45 nm in diameter). The genome consists of eleven double
stranded (ds) RNA segments. Both shells are composed of a single
polypeptide arranged in an icosahedral T= 13 arrangement (780
subunits). Sixty spike-like protrusions extend from the inner shell
through the outer one.
HIV, human immunodeficiency virus (Retroviridae): Spherical,
enveloped i.e. membrane bound particles (100-130 nm i diameter). The
genome consists of two (diploid) identical ss-RNA strands (plus t-RNA
from the host). As seen by thin-sectioning-techniques, the envelope
of immature virions is lined with a 20-25 nm thick submembraneous
layer of precursor (gag) proteins and the central area looks empty.
Mature virions have in the center a characteristic conical core
consisting of the viral RNA complexed with viral protein and the
submembraneous layer is very thin. Platina-carbon shadowing
techniques have revealed globular knobs (12-14 nm in diameter and 10
nm high) formed by clusters of glycoproteins. The knobs have the
tendency to come off; believed to make the viruses more resistant to
immunological attacks. However, the knobs are also sensitive to
preparative conditions. They are not easily distinguished e.g. in
thin sections. In negative-staining with uranyl acetate, merely
triangular shaped protrusions are detectable, interpreted that the
lower part of the knobs is triangular while the top is globular and
covered by stain i.e. making them less visible [17].
Our first reconstructions of early and late budding Uukuniemi virus
were done from sectioned material and showed truncated i.e. half
viruses indicating the inconvenience of using sections. Also
tilt-series of negatively stained whole-mounted Uukuniemi virus and
rotavirus resulted in collapsed i.e. 50% flattened
reconstructions.
A rather successful reconstruction of an Uukuniemi virus could be
obtained by staining the virus briefly with uranyl acetate in
methanol and sublimation from solidified t-butanol (Fig. 4). The
tomography could be determined by low-pass filtering down to 3 nm.
The shape of the reconstructed virus (71 nm i diameter) was well
preserved. Inside the particle we can also vaguely detect domains of
4 nm thick coiled strands, which could represent the core of
nucleoproteins. However, the pattern of knobs on the envelope was
less apparent compared to ordinary negative-staining.
These discrepancies of earlier techniques obligated us to develop new
preservation methods which can be used for electron tomography of
whole-mounted viruses.
Some new preparation methods emerged which we have tested on
rotavirus as a model. Briefly, in the method we have combined uranyl
acetate staining of glutaraldehyde-ruthenium-red fixed viruses with
sublimation from the t-butanol. Ruthenium red, which shows and
preserves carbohydrate moieties (for references see [6]) usually in
combination with osmium tetraoxid, seem to also retain the uranyl
acetate stain as control preparations without ruthenium red appear to
have lost the positive uranyl acetate staining. The shape of the
rotavirus is well preserved (Fig. 5) together with the pattern of
protrusions in the shell that usually can only be detected with
negative-staining. According to our measurements from the tomography,
the particle is probably representing a single-shelled rotavirion as
it is not more than 42 nm in diameter. The burdock-like particle is
formed by an inner core surrounded by numerous 8 nm long spikes with
a 4 nm wide globular top part.
The membrane wrapped HIV preparations was fixed in
glutaraldehyde-ruthenium red, furthermore treated with a
osmium-thiocarbohydrazide method [3] and briefly stained in uranyl
acetate in methanol before sublimation from solidified t-butanol.
Tomographies of HIV (Fig. 6) show an enveloped virus of 86 nm in
diameter. Inside the envelope material is found which is not fully
developed to a typical conical shape, distinctive of mature HIV
particles.
We have observed, that the diameters of the reconstructed viruses,
but not their subunits, are smaller than have been reported by others
[15-17]. This could be due to that our results are based on veritable
preservation of the 3D-shape of the particles and measurements in 3D
as our electron tomography realizes.
Future scenarios
Reconstruction of whole animal cells (about 10-30 mm in diameter)
must be done from serial sections as cells are too thick for ordinary
100 kV electron microscopes, which can only use sections not thicker
than about 0.2 mm. This will result in some 100 sections. For a 400
kV electron microscope the sections can be 3 mm thick, which would
result in not more than about 10 sections. As the information is in
the sections, a reconstruction of every section has to be completed
separately, with the necessary resolution needed, and merged into a
complete cell.
Perhaps, before the end of the century, we can experience when some
bionauts, ready with 3D-helmets, -gloves, -cameras and other
additional outfits, are entering the virtual world where a first
complete reconstruction of an entire eukaryotic cell is floating. The
reconstructed cell has been aligned from several serial sections,
each of which has been separately reconstructed at high resolution.
The whole reconstruction consists of a package of several terabytes
of voxels. All other activities for the computer have been shut off
for several hours so the computer can load the reconstructed volume.
The first voyage into an animal cell can begin. The scenario can take
place at CSC, who knows?
References
[1] Frank J, ed. Electron Tomography: Three-Dimensional Imaging
with the Transmission Electron Microscope. New York: Plenum Press,
1992.
[2] Skoglund U and Daneholt, B. Electron microscope tomography. TIBS
1986;11: 499-503.
[3] Takayama S and Hiramatsu H. Scanning electron microscopy of
centromeric region of L-cell chromosomes after treatment with Hoechst
33258 combined with 5-bromodeoxyuri dine. Chromosoma 1993;
102:227-232.
[4] Luther PK. Sample shrinkage and radiation. In: Frank J, ed.
Electron Tomography: Three-Dimensional Imaging with the Transmission
Electron Microscope. New York: Plenum Press, 1992: 39-60.
[5] Woodcock CL. The organization of chromosomes and chromatin. In:
Frank J, ed. Electron Tomography: Three-Dimensional Imaging with the
Transmission Electron Microscope. New York: Plenum Press, 1992:
313-357.
[6]Engelhardt
P. Higher Order Structure of Eukaryotic Chromosomes - Scaffolding
elements and DNA folding (Doctoral Dissertation). University of
Helsinki, 1988, ISBN 952-90032-1-8.
[7] Dierksen K Typke, D., Hegerl, R., Koster, A. J. and Baumeister,
W. Towards auto matic electron tomography. Ultramicroscopy 1992;
40:71-87.
[8] Koster AJ Chen, H., Sedat, J. W., and Agard, D. A. Automated
microscopy for electron tomography. Ultramicroscopy 1992;
46:207-27.
[9] Dierksen K Typke, D., Hegerl, R., and Baumeister, W. Towards
automatic electron tomography II. Implementation of autofocus and
low-dose procedures. Ultramicroscopy 1993; 49:109-120.
[10] Engelhardt P. 3D image processing of whole mounted chromosomes
by computer animation from tilted serial electron microscopic
projections. Conf. Proc: 3D Imaging Sciences in Microscopy.
Amsterdam: The Society for 3-D Imaging Sciences in Microscopy,
Amsterdam, 1992.
[11] Dolenc A. An overview of Rapid Prototyping Technologies in
manufacturing. In: Technical Report TKO-B116, Helsinki University of
Technology, April, 1994.
[12] Engelhardt P Ruokolainen, J., Dolenc, A., Öfverstedt, L-G.,
Mehlin, H. and Skoglund, U. 3D-reconstruction by
electron
tomography (EMT) of whole-mounted DNA-depleted metaphase
chromosomes show scaffolding macro coils, 30-nm fibers and 30-nm
particles. Conf. Proc: 3-D Image Processing in Microscopy. Munich:
The Society for 3-D Imaging Sciences in Microscopy, Amsterdam,
1994.
[13] Ris H. Higher order structures in chromosomes. 9th International
Congress on Electron Microscopy. Toronto: Microscopical Society of
Canada, 1978: 545-556.
[14] Engelhardt P Plagens, U., Zbarsky, I. B., and Filatova, L. S.
Granules 25-30 nm in di ameter: Basic constitutes of the nuclear
matrix, chromosome scaffold, and nuclear envelope. Proc. Natl. Acad.
Sci. U.S.A. 1982; 79:6937-6940.
[15] Petterson R and von Bonsdorff, C-H. Bunyaviridae. In: Nermut MV
Steven, A. C., eds. Animal Virus Structure. Elsevier Science
Publishers B. V. (Biomedical Division), 1987: 147-157.
[16] Prasad BVV Wang, G. J., Clerx, J. M. M., and Chiu, W.
Three-dimensional structure of rotavirus. J. Mol. Biol.
1988;199:269-275.
[17] Nermut MV. Electron microscopy of human immunodeficiency virus.
Microscopy and Analysis 1994; July(30): 13-15.
Thanks for the visit. You are visitor number since Febrary 23 1997
since Febrary 23 1997
Peter Engelhardt / 1997
Email: Peter.Engelhardt@Helsinki.Fi
Available at
http://www.csc.fi/jpr/emt/engelhar/csc/CSC-94.html