Engelhardt P and Ruokolainen J*
Department of Virology, PO Box 21, Haartman Institute, FIN-00014
University of Helsinki, Finland, e-mail:
peter.engelhardt@helsinki.fi
*CSC-Scientific Computing, PO Box 405, FIN-02101 Espoo, Finland,
e-mail: jpr@csc.fi
Presented at EUREM '96, Electron tomography session B2, 26-30th August, Dublin
We have developed fast and extensively automated electron tomography programs for Silicon Graphics machines (1, 2). For 3D-reconstructions of chromosomes, HeLa-cells were collected by shaking after 12-48h with colchicine, and chromosomes isolated after (15') hypotony in (75 mM) KCl, washed with (60%) AA, with further boiling (0.5-2') in (50%) AA after application of the chromosomes by centrifugation (2000 rpm, 2') on to fiducial gold-marked polystyrene-coated-Ni-EM-grids. The preparations were dehydrated in methanol, stained (30'') in (0.01-0.02%) uranyl acetate in methanol and freeze-dried from t-butanol (1). Tilt series (41 pictures, from 0 to + 60 degrees, every 3 degree) of the chromosomes were taken at 19000-58000x magnification with 100 kW in Jeol 100CX2 EM. The negatives were digitalized on a flat bed scanner (Microtek III, Agfa Arcus II, at 508, 1016 or 1200 dpi).
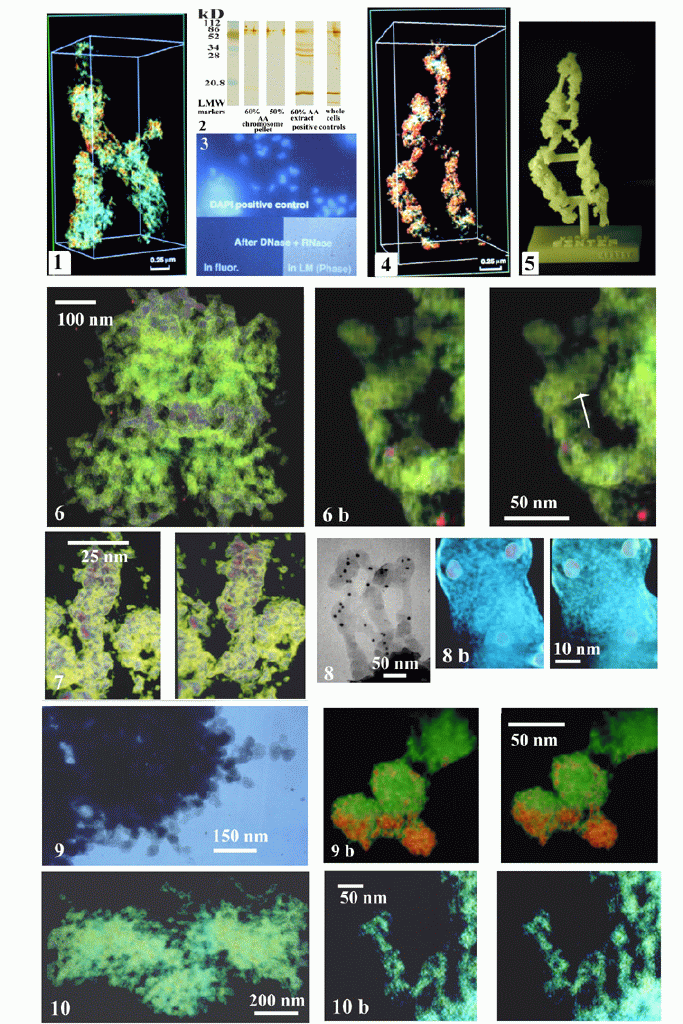
Chromosomes show excellent gross and fine structural morphology e.g. chromosomal macro coils and 30-nm thick chromatin fibers in 3D-reconstruction (Fig. 1), though depleted of most histones, major part of which are found in the (60%) AA extract, as shown with antibodies to H1 and H2b on immunoblottings (Fig. 2). Furthermore, digestion with nucleases (DNase I and RNase A) remove all detectable DNA-specific-fluorescence (Fig. 3), whereas controls show intensive staining with DNA-fluorochromes (DAPI). 3D-reconstruction of histone and DNA-depleted chromosomes i.e. chromosome scaffolds show macro-coiling and 30-nm thick scaffold fibers demonstrating that chromosome coiling is due to scaffolding proteins (Fig. 4). The chromosome scaffold can also be studied in detail on a solid-3D-plastic-model (Fig. 5) produced by "Rapid Prototyping Technology" methods that precisely 3D-reproduce the original reconstruction (1).
Our maximum entropy method, compared to the weighted back-projection method, shows superior quality i.e. practically noise free 3D-reconstructions of whole-mounted chromosomes and chromosome scaffolds (Fig. 6) that expose substructural details of the 30-nm chromosome and 30-nm scaffold fibers. With DNA present, the chromosome fibers look dense (Fig.7) with decorations that are different from the pattern seen in 30-nm scaffold fibers (Fig. 6b) i.e. depleted of DNA. The pattern in the scaffold fibers are seen as composed of a spiral with peripherally located toroid-like structures of about 8-nm in outer diameter with a central hole of about 4-nm, but also open-toroids are noticed. At high resolution we can detect as if a thin peripheral lace (3.5-nm outer, 1.5-nm inner mesh diameter, 1.5-nm thick) is covering and constituting the scaffold fibers (Fig. 8, 8b). Using modification of shrinkage avoiding fixations with tannic acid (2) for the chromosomes we could observe that the mean diameter of the 30-nm chromatin fiber increased to 30-60-nm, exposing 30-60-nm globular domains of the chromatin fibers with demarcated decorations (Fig. 9). Also the scaffold fibers show variable 30-60-nm thickness appearing more skeletonized showing 10-nm internal empty spaces surrounded by 4-8-nm thick curling fibrous structures (Fig. 10, 10b).
Because of morphological similarity and similarity in the folding pattern of chromatin and scaffold fibers as shown with our AA method, we presume their close and integrated association. We suggest that the bundles of DNA-nucleosomes are integrated with the scaffolding fiber i.e. intercalated by topoisomerase II molecules, apparent as 8-nm toroidal structures (above), and other associations with scaffolding proteins (above). The earlier finding (3) of looping DNA from a centrally positioned scaffold is obviously an artefact due to salt extraction and water-spreading that obscurely separates the original cohesion.
FIGURES
REFERENCES
1. Engelhardt P, Ruokolainen J, Dolenc A (1994) CSC News 6 (4): 14-2
2.Ruokolainen J. and Engelhardt P. (1996) Conf: 3D Imaging Sciences in Microscopy, Oxford/
3. Paulson JR, Laemmli UK, (1977) Cell 12: 817-82
MOVIES
The chromosome in Fig.6 as a movie:
 Click here.
Click here.
The size of the movie is 336 Kb
1996 / Peter Engelhardt
peter.engelhardt@helsinki.fi