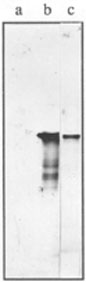
Fig. 1.
Immunoblotting of total proteins from E. coli.- a. 30 µg of total protein from Z905 recA) strain;
- b. 20 µg of total protein from AB1157 (recA+) strain;
- c. 5 ng of purified RecA protein of E. coli.
World Wide Web
available at http://www.csc.fi/jpr/emt/engelhar/ASCB.html
ATP-dependent homologous synaptase, or RecA protein, is a central enzyme of homologous recombination in procaryotes, facilitating both main reactions of the process, i. e. the initial homologous pairing and the strand exchange (for review, see [31]). This relatively small enzyme (close to 38-kDa), coded by the recA gene, has been described for more than 18 bacterial genera (for review, see [32]). All modern ideas of general recombination are based on the properties of RecA proteins. The search for a homologous recombinase in eucaryotes has been based on assays developed for testing the synaptase activity of RecA protein. Proteins with such activity have been discovered in fungi [23, 24, 35], plants [19] and in mammals, including transformed human cell lines [1, 5, 12, 19, 20, 22, 26, 29].
Interestingly, these proteins fall into two groups according to their requirement for ATP in the strand exchange reaction, i. e. ATP-dependent and ATP-independent, referred to as "m-rec" and "mAi-rec" proteins respectively. The enzymes of both types have been described in murine meiotic cells [17]. Both "m-rec" and "mAi-rec" proteins rise sharply during the early prophase reaching a peak at pachytene. However, whereas the activity of "m-rec" drops at the end of pachytene, the mAi-rec activity persists during the postmeiotic phase of spermatid development, and is also found in mature spermatozoa. The other important difference concerns the reaction with polyclonal antibodies to the E. coli RecA protein (anti-RecA), which bind to mAi-rec, inhibiting its activity, but do not react with the m-rec protein.
We speculate that different meiotic RecA-like proteins provide homologous recognition and pairing at different levels, molecular and cytological. Some observations presented in this work partially support this statement. Making use of the anti-RecA to analyze the pachytene nuclei isolated from bull testes, we detected staining of chromosome core structures with brightly staining nodules. This suggests the presence of RecA-like proteins in the structure of synaptonemal complexes (SCs) and in the recombination nodules. In immunoblotting, anti-RecA reacted strongly with a 40-kDa polypeptide and, more weakly, with 35- and 90-kDa polypeptides from pachytene nuclei.
Strains of E. coli K-12
AB1157 (relevant genotype recA+) and Z905 (relevant genotype as AB1157
but recA) [38] were used as a source of lysates and bacterial cell
proteins.
Isolation of pachytene nuclei
Pachytene nuclei were isolated from bull testes essentially according
to Hotta and Stern [18]. Isolated nuclei stored frozen (-70 oC or -20 oC) in 50% glycerin in TC buffer (10 mM Tris-HCl, pH 7.4; 3 mM CaCl2)
until used.
Purification of the E. coli RecA protein and preparation of polyclonal
antibodies (anti-RecA)
E. coli RecA protein was purified as described [6]. For immunisation
of rabbits, purified RecA protein was mixed with heat-denatured calf
thymus DNA (1 mole of RecA protein per 4 moles of nucleotides) in TN
buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl) and an equal volume of
complete Freund's adjuvant was added to the mixture. A total of 50 µg of
RecA protein was injected subcutaneously at one time. Subsequently, at
intervals of 7 days after the primary injection, three booster
injections were given using incomplete adjuvant. After one month of rest
the injection course was repeated. Seven days after the last booster the
rabbit was bled through the ear vein, and the presence of antibodies
against RecA protein was checked by dot-immunoblotting. Antibodies were
purified by ammonium sulfate precipitation and subsequent DEAE-cellulose
chromatography [14]. Finally they were absorbed with E. coli Z905
recA) lysates, to avoid nonspecific binding as described [33].
Affinity-purified antibodies against histone H1 (anti-H1) were a
generous gift from Dr. K. Khrapko [30].
Electrophoresis and immunoblotting.
Slab SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on
linear 7-16% gradients [25]. Proteins were then electrotransferred to
nitrocellulose membranes (BA-85, Schleicher & Schüll, Dassel, FRG) using
the procedure of Towbin et al. [37]. The filters were first stained with
Ponceau S in 3% trichloroacetic acid [14] to verify the transfer and to
aid in cutting each sample into strips. Then they were washed with TENT
buffer (50 mM Tris/HCl, pH 7.0, 5.0 mM EDTA, 150 mM NaCl, and 0.05%
(v/v) Tween-20) for 1 h and incubated with anti-RecA (diluted 1:600 in
TENT buffer) overnight at room temperature. After washing (4x10 min)
with TENT buffer they were incubated with swine anti-rabbit IgG
conjugated to peroxidase (DAKO, Copenhagen, Denmark) (diluted 1:200 in
TENT buffer) for 1 h at 37oC and washed (4x10 min) with TENT buffer.
After a final wash with TN buffer, the blots were stained for peroxidase
activity using TN buffer containing 0.5 mg/ml diaminobenzidine
hydrochloride (Sigma, St. Louis, MO) and fresh 0.1% (v/v) H2O2.
Indirect immunofluorescence microscopy of bull meiotic nuclei
Isolated nuclei, stored frozen, were diluted in TC buffer and attached
to freshly cleaved mica without any fixation [10]. Specimens were
incubated with anti-RecA (diluted 1:200-1:500 with Dulbecco's phoshpate
buffered saline (DPBS) containing Ca++, Mg++ and 0.2% or 0.5% bovine
serum albumin) for 1 h at room temperature. After washing with the
Dulbecco's solution they were incubated with fluorescein isothiocyanate
(FITC)-conjugated secondary antibodies, swine or goat anti-rabbit
immunoglobulins (Wellcome, Beckenham, UK) in the same solution (diluted
1:40) for 1 h at room temperature, washed and embedded in an anti-fading
solution [90% glycerin (Merck, glycerol for fluorescence microscopy) 40
mM Tris/HCl, pH 8.0, 3 mM CaCl2, 50-70 mM ß-mercaptoethanol with or
without 0.5-1 µg/ml propidium iodide]. All solutions contained 0.02%
NaN3 and were filtered through a 0.2 µm filter. The FITC-conjugated
secondary antibody solution was additionally pre-centrifuged in a
minifuge for 10-20 min. (+ 4-8 oC) at 14 000 rpm.
Treatments
The following treatments were done singly or in sequence on isolated
meiotic nuclei mounted on mica, after which they were processed as
above:
Detergent: In 0.2% Triton X-100 in Dulbecco's solution with several
changes from 10-20 min.
2 M salt extraction: In 2 M NaCl in TM buffer (10 mM Tris-HCL, pH 7.4; 5
mM MgCl2) from 10-30 min. with several changes and washed in TM buffer.
DNase I digestion: In 20-200 U/ml (RNase-free) DNase I, (Worthington or
Boehringer) in TM buffer for 1 h at room temperature and washed in TM
buffer.
Specimens were examined and photographed using an Olympus microscope with epifluorescent illumination. The following photographic films material were used: Fuji, p1600D chrome; Kodak (Ecktachrome p800/1600) Dia film; and Kodak, Ektapress gold 1600 color negative film.
The specificity of anti-RecA polyclonal antibodies was tested by immunoblotting of SDS-PAGE-separated samples of the E. coli strains AB1157 (recA+) and Z905 recA) as described in Material and Methods. A single major polypeptide band (close to 38-kDa) was recognized specifically by anti-RecA in recA+ cell extracts (Fig. 1, lane b). This band comigrated with purified E. coli RecA protein (close to 38-kDa). In some cell preparations anti-RecA recognized a few additional minor bands of low molecular weight (from 20- to 30-kDa). These polypeptides may result from a partial degradation of RecA protein and have been noted in similar experiments earlier [2]. By contrast, there were no cross-reactions of anti-RecA with any proteins of cell extracts prepared from bacterial cells bearing a recA gene deletion (Fig. 1, lane a).

Fig. 1.
Immunoblotting of total proteins from E. coli.
- a. 30 µg of total protein from Z905 recA) strain;
- b. 20 µg of total protein from AB1157 (recA+) strain;
- c. 5 ng of purified RecA protein of E. coli.
The intranuclear localization of antigens recognized by anti-RecA was studied in unfixed isolated bull meiotic nuclei by the indirect immunofluorescence technique. The pattern of antigen distribution was different in somatic and meiotic cell nuclei. In some nuclei of Sertoli cells, staining of nucleoli by anti-RecA could be recognized (Fig. 2a). Other somatic or premeiotic nuclei were stained only diffusely or insignificantly (Fig. 2b). However, in meiotic nuclei, anti-RecA stained distinct structures (Figs. 2a-d).
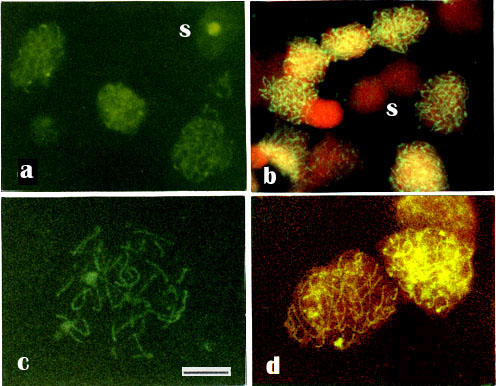
Fig. 2.
Indirect immunofluorescence of isolated unfixed nuclei from bull testes attached to freshly cleaved mica, using anti-RecA and FITC-conjugated secondary antibodies.
-a. Nuclei of Sertoli cells, showing staining of nucleoli.
-b. Other somatic and/or premeiotic nuclei (S), showing only diffuse or negligible FITC-staining, but primarily a red DNA staining with (1 µg/ml) propidium iodide (PI) counterstaining.
- c. Some late pachytene nuclei showing uniformly FITC-stained SCs
- d. The delicate green-stained SCs are surrounded by faint red-stained DNA in counterstaining with (0.5 µg/ml ) PI. Note the difference in staining pattern of meiotic and somatic nuclei. In pachytene nuclei distinctly stained fine meiotic chromosomal core structures can be seen, that we suggest are the synaptonemal complexes (SCs). Staining of somatic cell nuclei (S) are less prominent. - Bar 10 µm.
Pachytene nuclei showed staining of
fine meiotic chromosome cores, that we suggest are the elements of
synaptonemal complexes (SC) (Figs. 2a-d; 3a-e). This suggestion is
supported by the observation that the delicate green-stained cores were
surrounded by faintly red-stained DNA in counterstaining with propidium
iodide (Figs. 2b, d; 3b-e). In some nuclei, fine dots or nodules could
be seen on the thin cores, either along the cores (Figs. 3a, b, d, e,
and details at higher magnifications in insets), or at the telomeric
position (Fig. 3c and insets). In some favorably situated chromosome
cores (framed in Fig. 3) we could observe at higher magnifications that
some nodules could be described as ellipsoidal and some looked spherical
(seen as insets in Fig. 3). These structures could represent the
recombination nodules of early to late pachytene nuclei [4].
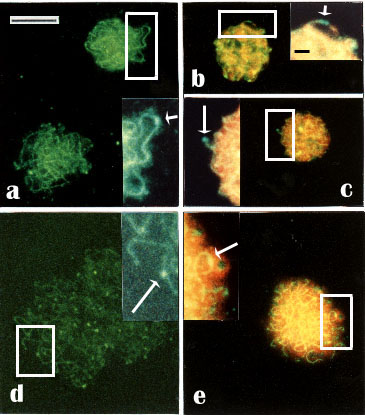
Fig. 3.
Indirect immunofluorescence using anti-RecA and FITC-conjugated secondary antibodies of meiotic nuclei with suggested recombination nodules. The nuclei represent early to late pachytene estimated by the presence of the SCs and the comparative size of the nuclei.
- a-c. Early to middle pachytene nuclei.
- d, e. Middle to late pachytene nuclei.
- a, d. PI omitted.
- b, c and -e. Counterstaining with PI.
- Insets: Details of the nodules (arrows) on the SCs indicate
- a, e. Ellipsoidal nodules.
- b, c and -d. Spherical nodules
- c. A terminal spherical nodule.
- b.Two spherical tandem nodules. Note, that many of the nuclei contain several nodules. Some chromosome cores with nodules (framed) are seen at higher magnification in the insets. - Bar 10 µm - In insets: -Bar 1 µm.
Spermatids and spermatozoa also show fluorescence with anti-RecA. A collar surrounding the dorsal end of the sperm head was usually seen, but also a sponge-like or granular-like staining at the top end of the head (Fig. 4a, b), suggesting that sperms carry RecA-related proteins.
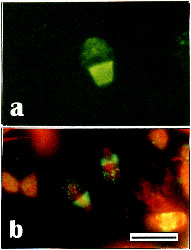
Fig. 4.
Distribution of antigens recognized by anti-RecA in bull spermatozoa without (a) or with (b) counterstaining with PI. Staining is prominent in the collar surrounding the dorsal end of the sperm-head but also a slight spongy-like or granular-like staining at the top end of the head can be seen. - Bar 10 µm.
Anti-RecA staining of meiotic nuclei was clearly specific. Incubation of nuclear preparations with preimmune serum produced only weak and diffuse FITC-staining; only reddish-stained nuclei were seen by PI (Fig. 5a, b). Furthermore, no positive staining could be detected after incubation of specimens with secondary antibodies alone (data not shown).
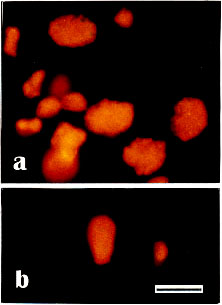
Fig. 5.
- a, b Control preparations of isolated bull meiotic nuclei and sperm-head incubated with preimmune serum. Preparations were counterstained with PI (0.5-1 µg/ml). No specific staining of chromosome structures, only homogeneous reddish PI-staining of DNA. - Bar 10 µm.
Pretreatment of nuclear preparations with 0.2% Triton X-100 and 2 M NaCl decreased slightly the staining of observed nuclear structures (Figs. 6a-c). However, after DNase digestion, the stained chromosome cores appeared smeared (Fig. 6d) and after additional 2 M salt extraction a halo surrounded a diffusely stained nuclei (Fig. 6e). Staining with propidium iodide was negative, demonstrating that all detectable DNA had been digested (compare Figs. 6b, c and 6d, e)
It should be noted that distinct structures were revealed by anti-RecA only on unfixed meiotic nuclei. After fixation of the nuclear preparation, anti-RecA stained them more diffusely (data not shown).
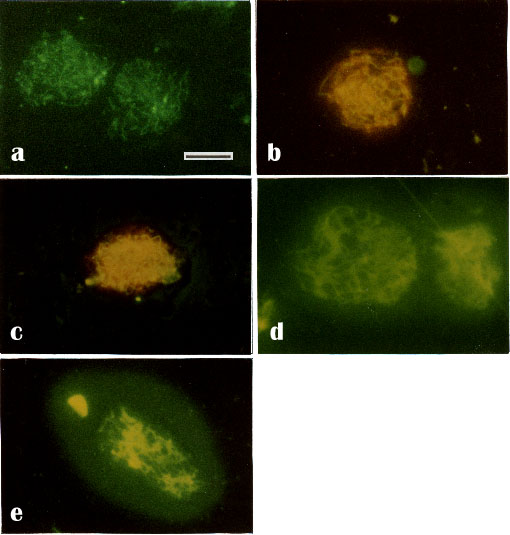
Fig. 6.
Immunofluorescence staining with anti-RecA of unfixed bull pachytene nuclei after treatment with: 2 M salt solution
(a) and (b, counterstained with PI) or 0.2% Triton X-100 and 2M salt solution
(c, counterstained with PI) or 0.2% Triton X-100 and DNase I (20-200 U/ml)
(d, counterstained with PI) or 0.2% Triton X-100, 2 M salt solution and DNase I (e, counterstained with PI). No red PI-staining of DNA can be detected (d, e) demonstrating that all detectable DNA has been digested. - Bar 10 µm.
In addition to immunofluorescence microscopy, we performed immunoblot analysis of proteins from isolated bull pachytene nuclei. As shown in Figure 7 anti-RecA reacted strongly with a polypeptide of approximately 40 kDa and, more weakly, with a 35- and a 90-kDa polypeptide. In addition, anti-RecA stained a broad lower molecular weight (15-20 kDa) zone, but this reaction was nonspecific since it was also recognized by anti-H1 (Fig. 7, lane e). However, anti-H1 did not cross-react with the 40- or 90-kDa polypeptides.
These 40- and 90-kD polypeptides were retained after treatment of pachytene nuclei with Triton X-100 (0.2%) or after DNase I (200 U/ml) digestion followed (in both cases) by NaCl (2 M) extraction (Fig. 7, lanes b, c); in the latter case the level of the polypeptides was slightly reduced.
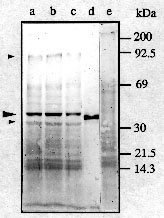
Fig. 7.
Immunoblotting of proteins from isolated bull pachytene nuclei.
- a, e. isolated untreated nuclei;
- b. nuclei treated with 0.2% Triton X-100;
- c. nuclei treated with 0.2% Triton X-100, DNase I (200 U/ml) and 2 M salt solution;
- d. 5 ng of purified RecA protein of E. coli. Blots were incubated with anti-RecA (a-d) or with anti-H1 (e).
Arrow heads indicate positions of polypeptides recognized specifically by anti-RecA.
The immunoreactive polypeptides demonstrated above differ markedly in their molecular weights from the 120-kDa KIN protein(s) detected in different mammalian somatic cell lines by polyclonal antibodies against E. coli RecA protein and which are essentially concentrated in the nucleus [2]. It has been shown earlier that these antibodies cross-reacted also with yeast ribonucleotide reductase [3, 8, 21]. The mammalian analogue of the enzyme is composed of two monomers of 45- and 88-kDa [36] values close to the molecular weights of the bull meiotic proteins described in the present report. However, unlike the latter, mammalian ribonucleotide reductase is located in the cytoplasm and not in the nucleus [11].
Our observations permit us to suggest the presence of RecA-like proteins in the composite structure of SC. We found one major and two minor polypeptides, based on their cross-reaction with anti-RecA. They differ in their molecular weights as well as in their localizations from components of synaptonemal complexes known so far. It has been shown that monoclonal antibodies against isolated rat SC recognized two polypeptides of 30 and 33 kDa localized in meiotic chromosome cores [15, 16, 28]. Moreover, unlike proteins described in the present paper, none of these antigens could be detected in spermatids or spermatozoa. We do not know of any earlier reports where nodules juxtaposed on the SCs can be detected in staining with antibodies, though many antibodies have been reported to stain the SCs (for review, see [7]). For instance, polyclonal antibodies to topoisomerase II, one of the constituents of the mitotic metaphase chromosome scaffold, stained chromosome cores in pachytene nuclei, but not recombination nodules [27].
We further suggest that these nodules, from their distribution in meiosis, their localizations on the SC and their anti-RecA positive nature, may represent the recombination nodules, and more specifically the late recombination nodules according to the specifications [4]) of early to late pachytene nuclei. We also observed nuclei (data not shown) with rather many nodules without any detectable chromosomal cores. These nodules could represent the early recombination nodules of earlier meiotic stages as described in electron microscopic investigations [4]. By contrast, uniformly stained cores must represent some late pachytene nuclei without nodules, or where the nodules are not recognized. This could be due to the preparative conditions, where the compression of nuclei may have stretched the chromosome cores so that the nodules are unrecognizable. This situation has also been recognized in the spreading of meiotic chromosomes for electron microscopic analyses [4].
Practically nothing is known about the composition of the recombination nodules, but various pieces of evidence indicate that recA-like proteins are involved in meiotic recombination (for review, see [34]). We could distinguish a few ellipsoidal and spherical nodules as they appear at higher magnifications (Insets in Fig. 3). It could be argued that they are too small to be resolved at the light microscopic level. So far, ellipsoidal and spherical recombination nodules have only been described with the electron microscope (for review, see [4]). However, this remark may not concern fluorescence microscopy as the glow effect caused by the fluorescence would significantly magnify these details. We can argue further that the thickness of the SC is compatible with dimensions of the recombination nodules, which could thus be visualized in fluorescence microscopy by analogy with the SCs (for details, see [4, 7, 13]).
Our observations suggest association of the anti-RecA staining antigens with DNA, since following digestion of DNA, the chromosome cores looked blurred, and dispersion of the antigen was evident. It is known that the SC is not disrupted by nuclease digestion or by extraction with 2 M salt (for references, see [7, 13]) This would indicate that the antigen is associated with the DNA strands that are in close association with the SC. Furthermore, it is tempting to speculate that these strands could be interacting in a single-stranded form associated with RecA-like protein(s).
One of the proteins could be the mAi-recA protein mentioned in the Introduction that cross-reacts with polyclonal anti-RecA and has a molecular weight of 40-kDa [17]. Interestingly, this protein promoted ATP-indepedent homologous DNA pairing. It may thus resemble the 40-kDa bull polypeptide demonstrated here. Experiments are now in progress to check this suggestion as well as to clarify the nature of the other proteins reacting with anti-RecA.
[1] Akhmedov, A. T., E. A. Namsaraev, E. M. Zaitseva, E. N. Zaitsev, V.
A. Lanzov: Study of recombination activity in mammalian cell extracts
(in Russian). Biopolymers and Cell (USSR) 6, 38-45 (1990).
[2] Angulo, J. F., P. L. Moreau, R. Maunoury, J. Laporte, A. M. Hill, R.
Bertolotti, R. Devoret: KIN, a mammalian nuclear protein
immunologically related to E. coli RecA protein. Mutat. Res. 217,
123-134 (1989).
[3] Angulo, J. F., J. Schwencke, P. L. Moreau, E. Moustacchi, R.
Devoret: A yeast protein analogous to E. coli RecA protein whose
cellular level is enhanced after UV-irradiation. Mol. Gen. Genet. 20,
20-124 (1985).
[4] Carpenter, A. T. C.: Thoughts on recombination nodules, meiotic
recombination, and chiasmata. In: R. Kucherlapati and G. R. Smith
(eds.): Genetic recombination. pp. 529-548. Amer. Soc. for Microbiol.
Washington, D. C. 1988.
[5] Cassuto, E., L. A. Lightfoot, P. Howard-Flanders: Partial
purification of an activity from human cells that promotes homologous
pairing and the formation of heteroduplex DNA in the presence of
ATP. Mol. Gen. Genet. 208, 10-14 (1987).
[6] Cox, M. M., K. McEntee, I. R. Lehman: A simple and rapid procedure
for the large scale purification of the recA protein of Escherichia
coli. J. Biol. Chem. 256, 4676-4678 (1981).
[7] Dresser, M. E.: The synaptonemal complex and meiosis: An
immunochemical approach. In: P. B. Moens (ed.): Meiosis. pp. 245-274.
Academic Press. New York 1987.
[8] Elledge, S., R. Davis: Identification and isolation of the gene
encoding the small subunit of ribonucleotide reductase from
Saccharomyces cerevisiae: DNA damage-inducible gene required for mitotic
viability. Mol. Cell. Biol. 7, 2783-2793 (1987).
[9] Engelhardt, P., A. T. Akhmedov, V. A. Lanzov: Anti-recA recognize
chromosome cores and nodules in meiotic nuclei. J. Cell. Biol. 111, 376a
(1990).
[10] Engelhardt, P., U. Plagens, I. B. Zbarsky, L. S. Filatova: Granules
25-30 nm in diameter: Basic constituent of the nuclear matrix,
chromosome scaf?fold, and nuclear envelope. Proc. Natl. Acad. Sci. USA
79, 6937-6940 (1982).
[11] Engstrńm, Y., B. Rozell, H. A. Hansson, S. Stemme, L. Thelander:
Localization of ribonucleotide reductase in mammalian cells. EMBO. J. 3,
863-867 (1984).
[12] Ganea, D., P. Moore, L. Chekuri, R. Kucherlapati: Characterization
of an ATP-dependent DNA strand transferase from human cells. Mol.
Cell. Biol. 7, 3124-3130 (1987).
[13] Giroux, C. N.: Chromosome synapsis and meiotic recombination. In:
R. Kucherlapati and G. R. Smith (eds.): Genetic recombination. pp.
465-496. Amer. Soc. for Microbiol. Washington, D. C. 1988.
[14] Harlow, E., D. Lane: Antibodies: A Laboratory Manual. Cold Spring
Harbor Laboratory Press. New York 1988.
[15] Heyting, C., R. J. Dettmers, A. J. Dietrich, E. J. Redeker, A. C.
Vink: Two major components of synaptonemal complexes are specific
for meiotic prophase nuclei. Chromosoma 96, 325-332 (1988).
[16] Heyting, C., P. B. Moens, W. van Raamsdonk, A. J. Dietrich, A. C.
Vink, E. J. Redeker: Identification of two major components of the
lateral elements of synaptonemal complexes of the rat. Eur. J. Cell
Biol. 43, 148-154 (1987).
[17] Higashitani, A., S. Tabata, T. Ogawa, H. Ogawa, M. Shibata, Y.
Hotta: ATP-independent strand transfer protein from murine
spermatocytes, spermatids, and spermatozoa. Exp. Cell Res. 186, 317-323
(1990).
[18] Hotta, Y., H. Stern: Meiotic protein in spermatocytes of mammals.
Nature New Biol. 234, 83-86 (1971).
[19] Hotta, Y., S. Tabata, R. A. Bouchard, R. Pinon, H. Stern: General
recombination mechanisms in extracts of meiotic cells. Chromosoma 93,
140-151 (1985).
[20] Hsieh, P., M. S. Meyn, R. D. Camerini-Otero: Partial purification
and characterization of a recombinase from human cells. Cell 44, 885-894
(1986).
[21] Hurd, H. K., C. W. Roberts, J. W. Roberts: Identification of the
gene for the yeast ribonucleotide reductase small subunit and its
inducibility by methyl methanesulfonate. Mol. Cell. Biol. 7, 3673-3677
(1987).
[22] Kenne, K., S. Ljungquist: RecA-like activity in mammalian cell
extracts of different origin. Mutation Res. 184, 229-236 (1987).
[23] Kmiec, E., W. K. Holloman: Homologous pairing of DNA molecules
promoted by a protein from Ustilago. Cell 29, 367-374 (1982).
[24] Kolodner, R., D. H. Evans, P. T. Morrison: Purification and
characterization of an activity from Saccharomyces cerevisiae that
catalyzes homologous pairing and strand exchange. Proc. Natl. Acad. Sci. USA 84, 5560-5564 (1987).
[25] Laemmli, U. K.: Cleavage of structural proteins during the
assembly of the head of bacteriophage T4. Nature 227, 680-685 (1970).
[26] Lopez, B., S. Rousset, J. Coppey: Homologous recombination
intermediates between two duplex DNA catalysed by human cell
extracts. Nucleic Acids Res. 15, 5643-5655 (1987).
[27] Moens, P. B., W. C. Earnshaw: Anti-topoisomerase II recognizes
meiotic chromosome cores. Chromosoma 98, 317-322 (1989).
[28] Moens, P. B., C. Heyting, A. J. Dietrich, W. van Raamsdonk, Q.
Chen: Synaptonemal complex antigen location and conservation. J. Cell
Biol. 105, 93-103 (1987).
[29] Moore, S. P., R. Fishel: Purification and characterization of a
protein from human cells which promotes homologous pairing of DNA. J.
Biol. Chem. 265, 11108-11117 (1990).
[30] Postnikov, Y. V., V. V. Shick, A. V. Belyavsky, K. R. Khrapko, K.
L. Brodolin, T. A. Nikolskaya, A. D. Mirzabekov: Distribution of high
mobility group proteins 1/2, E and 14/17 and linker histones H1 and H5
on transcribed and non-transcribed regions of chicken erythrocyte
chromatin. Nucleic Acids Res. 19, 717-725 (1991).
[31] Radding, C. M.: Helical RecA nucleoprotein filaments mediate
homologous pairing and strand exchange. Biochim. Biophys. Acta 1008,
131-145 (1989).
[32] Roca, A. I., M. M. Cox: The RecA protein: Structure and function.
Crit. Rev. Biochem. Molec. Biol. 25, 415-456 (1990).
[33] Sambrook, J., E. F. Fritsch, T. Maniatis: Molecular Cloning: A
Laboratory Manual, 2nd. ed. Cold Spring Harbor Laboratory Press. New
York 1989.
[34] Stern, H., Y. Hotta: The biochemistry of meiosis. In: P. B. Moens
(eds.): Meiosis. pp. 303-329. Academic Press. New York. 1987.
[35] Sugino, A., J. Nitiss, M. A. Resnick: ATP-independent DNA strand
transfer catalyzed by protein(s) from meiotic cells of the yeast
Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 85, 3683-3687
(1988).
[36] Thelander, L., P. Berg: Isolation and characterization of
expressible cDNA clones encoding the M1 and M2 subunits of mouse
ribonucleotide reductase. Mol. Cell. Biol. 6, 3433-3442 (1986).
[37] Towbin, H., T. Staehelin, J. Gordon: Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76, 4350-4354 (1979).
[38] Zaitsev, E. N., E. M. Zaitseva, I. V. Bakhlanova, V. N. Gorelov, N.
P. Kuzmin, V. M. Kryukov, V. A. Lanzov: Cloning and characteristics of
recA gene in Pseudomonas aeruginosa (in Russian). Genetika (USSR) 22,
2721-2727 (1986).